1630
The Role of Bias Field Correction in the Free Water Elimination Problem1Radiology, University of Pennsylvania, Philadelphia, PA, United States, 2Synaptive Medical Inc., Toronto, ON, Canada
Synopsis
Free water elimination (FWE) paradigms provide information about underlying pathology-induced tissue changes, based on a multi-compartment fit to the dMRI acquisition. Non-uniform intensity in MR signal, either due to coil or acquisition sequence, produces inhomogeneous tissue intensity profiles. This negatively affects FWE paradigms, producing artifactual multi-compartment fits. In this work, through extensive application on varied datasets, we demonstrate the effect of using bias field correction, an optimized non-uniform intensity normalization, on reducing artifacts in FWE and producing physiologically relevant maps. This suggests that bias correction should be maintained as an essential step in dMRI preprocessing for FWE.
Purpose
Accurate estimation of the free water map using FWE paradigms on dMRI acquisitions is a clinically significant problem because of the information free water maps potentially provide about pathologies like brain tumors and traumatic brain injury. Use of single shell dMRI sequences for FWE is the only clinically feasible option currently. However, the under-determined nature of FWE using single shell acquisition1, produces artifactual voxels with diffusivity values that disagree with the physical properties of the human brain, irrespective of the choice of parameters. More critically, these artifactual voxels have high FWE-FA values, which can be problematic for tractography and neurosurgical planning based on tract extraction. We hypothesize that this artifactual fit is related to inhomogeneity in the MR signal, and will be significantly alleviated by bias field correction of the data. With the aim of eliminating these artifacts, we propose that using N4ITK bias correction to obtain uniform intensity profiles for various tissue types is key to reducing these voxels in the FWE output. We demonstrate, across ten datasets of healthy controls and tumor patients, with various acquisition protocols, scanner and coil types, that bias correction can significantly lower the artifactual voxels in FWE paradigms, leading to plausible free water correction in edema and healthy white matter structures.Methods
1- Datasets:
A- Healthy Control Datasets:
Dataset1: 95 subjects from the Philadelphia Neurodevelopmental Cohort2 (TR/TE=8100/82ms, b=1000s/mm2, 64 weighted diffusion directions, and a spatial resolution of 1.875x1.875x2mm).
Dataset2: 95 subjects that served as controls for an autism spectrum disorder study3 (TR/TE=11,000/76ms, b=1000s/mm2, 30 diffusion gradients, and 2mm isotropic spatial resolution).
Dataset3&4: (ISMRM TraCED Challenge) Five acquisitions that were repeated in two sessions and acquired in two different scanners of a single subject. For this study, we used the data from the shell of b=1000 s/mm2, extracted from the 3-shell acquisition. Details about the challenge data are available in4.
Dataset5,6&7: Nine subjects scanned at 3 time points separated by 2 weeks5. (TR/TE=14800/111ms, b=1000s/mm2, 64 encoding diffusion gradients, and 2mm isotropic spatial resolution).
B- Tumor Datasets
Dataset8: 10 patients were selected from an ongoing tracking tumor study. These patients underwent a Multishell acquisition from which the b=800 s/mm2 shell was extracted (TR/TE=5216/100ms, 30 encoding diffusion gradients, and an isotropic spatial resolution of 2mm).
Dataset9: The same patients in Dataset8 underwent a single shell diffusion sequence. (TE=6300/100ms, b=1000s/mm2, 30 encoding diffusion gradients, and a spatial resolution of 1.71x1.71x3mm).
Dataset10: Ten patients, five glioblastoma multiforme (GBM) and 5 brain metastasis (TR/TE=5000/86ms, and b=1000s/mm2, and 30 weighted diffusion directions).
2- Bias Correction & FWE:
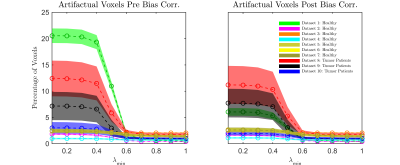
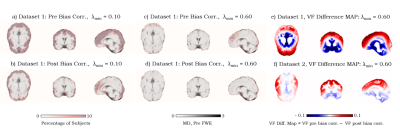
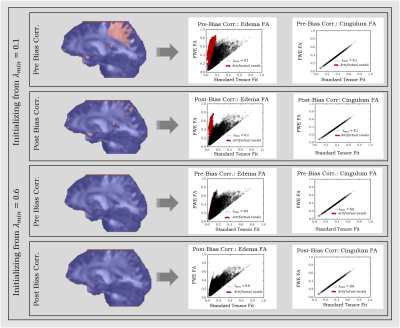
Datasets were bias corrected by employing N4ITK6. FWE was carried out1, by initializing the tensors with an algorithm-specific parameter λmin that varies from 0.1 x10-3mm2/s1 to 1.0 x10-3mm2/s and a fixed λmax of 2.5x10-3mm2/s1, representing the minimum and the maximum of the expected range of tissue diffusivities in the brain. Artifactual voxels were defined as voxels with MD<0.4 x10-3mm-3/s7. The percentages of these voxels in the brain mask were determined in all datasets across different λmin values (Fig.1). For large healthy datasets (1&2), subjects were registered to the same space, and the percentage of subjects that exhibit artifact at each voxel was calculated along with the difference in the volume fraction map between pre and post N4ITK (Fig.2). FWE-based FA was computed and compared to that from the standard tensor fit in both edema and cingulum masks.
Results and Discussion
Using N4ITK, with the proper choice of the initializing parameters (that match the physical properties of the brain tissue), dramatically reduces the percentage of artifactual voxels across all datasets (Fig.1). Despite the variation across different scanners in lower λmin values, all investigated protocols converge to λmin=0.6x10-3mm2/s as an optimal initializer for single shell dMRI-based FWE. Investigating large healthy control datasets (1&2) shows that the free water map is overestimated in cortical regions, where the bias in b0 signal profile is observed (Fig.2). To verify that eliminating the artifacts doesn’t come at the cost of estimating the free water content, FA maps of edema and cingulum masks were investigated. Our results demonstrate that FWE post bias correction provides a robust free water correction in the edema region while obtaining FA values in the contralateral cingulum (minimally affected by free water) that are consistent with those from the standard tensor fit (Fig.3).Conclusion
Across a large number of datasets, we demonstrated that bias correction alleviates significantly the artifactual voxels in FWE output, while maintaining a plausible solution. We also show that the same set of parameters of the FWE paradigm produce robust free water maps, post bias correction, despite their variation in scanners and acquisition parameters. This demonstrates the importance of bias correction in any dMRI processing involving FWE.Acknowledgements
This research was supported by the National Institutes of Health (NIH) grant 1R01NS096606 (PI: Ragini Verma), and research grant from Synaptive Medical 30071788 (PI: Ragini Verma).References
1- Pasternak, O., et al., Free water elimination and mapping from diffusion MRI. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine, 2009. 62: p. 717-30.
2- Satterthwaite, T.D., et al., The Philadelphia Neurodevelopmental Cohort: A publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage, 2016. 124(Pt B): p. 1115-9.
3- Ghanbari, Y., et al., Identifying group discriminative and age regressive sub-networks from DTI-based connectivity via a unified framework of non-negative matrix factorization and graph embedding. Med Image Anal, 2014. 18(8): p. 1337-48.
4- https://my.vanderbilt.edu/ismrmtraced2017/
5- Tunc, B., et al., Automated tract extraction via atlas based Adaptive Clustering. Neuroimage, 2014. 102P2: p. 596-607.
6- Tustison, N.J., et al., N4ITK: Improved N3 Bias Correction. IEEE Transactions on Medical Imaging, 2010. 29(6): p. 1310-1320.
7- Helenius, J., et al., Diffusion-Weighted MR Imaging in Normal Human Brains in Various Age Groups. AJNR Am J Neuroradiol, 2002. 23(2): p. 194-199.
Figures