1533
The Immune Checkpoint PD-L1 and Choline Kinase-α are inversely related in triple negative human breast cancer cells1Division of Cancer Imaging Research, The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
Immune checkpoint inhibition to activate the immune system has emerged as an exciting treatment option for several cancers. Programmed death-ligand 1 (PD-L1) plays a major role in immune suppression. We investigated the relationship between the aberrant choline metabolism observed in most cancers and PD-L1 expression in triple negative human MDA-MB-231 breast cancer cells. Using siRNA to downregulate choline kinase-α (Chk-α) or PD-L1 or both, we identified a close inverse interdependence between Chk-α, PD-L1 and phosphocholine. These results have significant implications for treatments that decrease Chk-α expression as these may drive up PD-L1 expression allowing escape of cancer cells from immune surveillance.
Introduction
Evading immune surveillance has emerged as a hallmark of cancer1. T-cells have “checkpoints” such as PD-1 (programmed death-1) to guard against autoimmunity and protect tissues from damage during strong response to infection 2,3. Cancer cells take advantage of these immune suppression mechanisms by expressing programmed cell death-1 ligand (PD-L1) in response to the interferon secreted by effector T-cells during an immune response 4,5. Once PD-L1 binds to PD-1 the T cell becomes deactivated. Checkpoint inhibitor therapies trigger an immune response against cancer cells by blocking immune checkpoints used by cancer cells to escape immune destruction. PD-L1 is an immune checkpoint overexpressed in cancers, including breast cancer, that has been successfully exploited for immune therapy6. Increased phosphocholine (PC), detected as an increase of total choline by 1H-MRS in most cancers, is mainly due to increased expression of choline kinase-a (Chk-α). Since the presence of PC on proteins has been related to escape from immune surveillance7, here, for the first time, we investigated the relationship between Chk-α, PC and PD-L1 in triple negative MDA-MB-231 human breast cancer cells.Methods
Experiments were performed using triple negative breast MDA-MB-231 cells that were transiently transfected with small interfering RNA (siRNA) against either luciferase (used as control siRNA), Chk-α or PD-L1 following standard protocols. Total RNA was isolated, complementary cDNA synthesised and quantitative real-time PCR (q-RT-PCR) performed using IQ SYBR Green supermix and gene specific primers following an established protocol8.
For high resolution 1H-MRS, cell extracts were obtained using a dual-phase extraction method as previously described8. Water-soluble samples were dissolved in 0.6 mL of buffered D2O (Sigma). High-resolution 1H-MR spectra were recorded on a Bruker Biospin Avance-III 750 MHz NMR (Bruker Biospin) spectrometer operating at a proton frequency of 750.21 MHz using a 5-mm broad band inverse (BBI) probe head equipped with z-gradient accessories. Zgpr pulse program was applied. 1H-MR spectra were manually phased and automated baseline corrected using TOPSPIN 3.2 software. Integrals of the metabolites of interest were determined and normalized to the TSP reference and the number of cells. Metabolites were estimated from at least three experimental samples. Statistical significance was evaluated using the Student t test.
Results and Discussion
As shown in Figure 1, silencing Chk-α resulted in a significant increase of PD-L1 mRNA expression and silencing PD-L1 resulted in a significant increase of Chk-α mRNA expression. This inverse relationship was eliminated when both PD-L1 and Chk-α were silenced. Transfection with control siRNA did not affect in Chk-α mRNA, but induced a small increase of PD-L1 mRNA compared to untreated cells. Transfection of cells with Chk-α and PD-L1 siRNA resulted in significant decreases of CHk-α and PD-L1 mRNA.
To demonstrate that changes in Chk-α mRNA following PD-L1 down regulation translated to functional changes, we carried out high resolution 1H-MRS of cell extracts to determine choline (Cho), PC, and glycerophosphocholine (GPC) levels, as shown in the representative spectra in Figures 2A-D. Consistent with the mRNA results, a significant increase of PC was observed in spectra obtained from cells transfected with PD-L1 siRNA compared to cells transfected with control siRNA (Figure 2E). Also consistent with the mRNA results, cells transfected with a combination of Chk-α and PD-L1 siRNA showed a significant decrease of PC (Figure 2E).
Our data have identified, for the first time, the inverse association between PD-L1 and Chk-α expression. These data suggest that treatments that decrease Chk-α and PC could result in cancer cells escaping immune surveillance through increased expression of PD-L1, but these cells may become more susceptible to checkpoint inhibitors such as anti-PD-L1 or anti-PD-1 antibodies. Our ongoing studies are characterizing the effects of anti-PD-L1 antibody treatment on Chk-α expression.
Acknowledgements
This work was supported by NIH R35CA209960 and R01CA82337, and the Emerson Foundation. JPT was supported by Fundacion Alsonso Martin Escudero.References
1. Hanahan D and Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144(5):646-74.
2. Shin DS and Ribas A. The evolution of checkpoint blockade as a cancer therapy: what's here, what's next? Curr Opin Immunol. 2015; 33:23-35.
3. Suzuki S, Ishida T, Yoshikawa K, et al. Current status of immunotherapy. Jpn J Clin Oncol. 2016.
4. Hirano F, Kaneko K, Tamura H, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005; 65(3):1089-96.
5. Homet Moreno B and Ribas A. Anti-programmed cell death protein-1/ligand-1 therapy in different cancers. Br J Cancer. 2015; 112(9):1421-7.
6. Mittendorf EA, Philips AV, Meric-Bernstam F, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014; 2(4):361-70.
7. Lovell TM, Woods RJ, Butlin DJ, et al. Identification of a novel mammalian post-translational modification, phosphocholine, on placental secretory polypeptides. J Mol Endocrinol. 2007; 39(3):189-98.
8. Penet MF, Shah T,
Bharti S, et al. Metabolic imaging of pancreatic ductal adenocarcinoma detects
altered choline metabolism. Clin Cancer Res. 2015; 21(2):386-95.
Figures
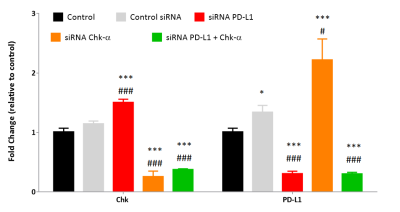
Figure 1. Relative fold change of choline kinase-α (Chk-α) and programmed cell death-1 ligand (PD-L1) mRNA expression in MDA-MB-231 cells: untreated (black), transfected with 100 nM control siRNA (gray), transfected with 100 nM PD-L1 siRNA (red), transfected with 100 nM Chk-α siRNA (orange), transfected with a mixture of 50 nM PD-L1 and 50 nM Chk-α (green) siRNA. Values represent Mean ± SEM from at least five independent experiments.
*p ≤ 0.05 **p ≤ 0.01 ***p ≤ 0.001 compared to untreated cells.
# p ≤ 0.05 ##p ≤ 0.01 ###p ≤ 0.001 compared to cells transfected with control siRNA.
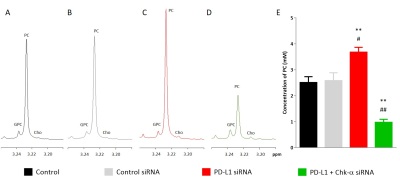
Figure 2. Representative 1H-MR spectra showing changes in glycerophosphocholine (GPC), phosphocholine (PC) and choline (Cho) in MDA-MB-231 cells: untreated (A, black), transfected with 100 nM control siRNA (B, gray), transfected with 100 nM PD-L1 siRNA (C, red), transfected with a mixture of 50 nM PD-L1 and 50 nM Chk-α siRNA (D, green). Quantification of PC in cell extracts (E). Values represent Mean ± SEM from at least three independent experiments.
*p ≤ 0.05 **p ≤ 0.01 compared to untreated cells.
#p ≤ 0.05 ##p ≤ 0.01 compared to cells treated with control siRNA.