1516
Automatic classification between high grade gliomas and brain metastasis using Bag-Of-Features in comparison to statistical and morphologic features1Functional Brain Center, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 2Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 3Department of Chemical Physics, Weizmann Institute, Rehovot, Israel, 4Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel
Synopsis
This study suggests a clinical decision-support tool for automatic classification of brain tumors. Classification was performed on 179 MRI patients: 81 patients with high grade-gliomas (HGG) and 98 patients with brain metastases (MET, 55 breast, 43 lung, cancer origin). The input data were Bag-Of-Features (BoF) and statistical-&-morphologic features extracted from T1WI+Gd. Classification was performed using five ensemble classifiers and results were evaluated using five-fold cross-validation. Best classification results produced accuracy=83%, sensitivity=87%, and specificity=81% for discriminating between HGG and MET using Statistical-&-morphologic features, and accuracy=79%, sensitivity=76%, and specificity=80% for discriminating between breast and lung MET using BoF + Statistical-&-morphologic features.
INTRODUCTION
High grade gliomas (HGG) and brain metastasis (MET) are the two most common malignant brain tumors in adults. There is high clinical importance for differentiating between the two, as medical staging, surgical planning, and therapeutic decisions are vastly different for each tumor type1,2. Multiplicity, location and morphology features can assist in differentiating HGG from MET. However despite its high sensitivity, the ability to differentiate HGG from single brain metastasis is frequently a challenge due to their similar imaging appearance on MRI3,4 with 11% of patients with a brain lesion given a false-positive diagnosis, based on conventional MRI only5,6. The aim of this study was to automatically differentiate high-grade gliomas from MET and breast from lung MET based on 1. Bag-Of-Features and 2. Statistical-&-morphologic features, extracted from post contrast T1 weighted images.METHODS
Patients: In this retrospective study 179 pre-operative MRI scans, obtained from newly diagnosed patients with brain tumors: 81 patients with HGG, and 98 patients with brain MET (55 from breast cancer and 43 from lung cancer origin), all proven by histopathology.
MRI data: included 3D post contrast T1WI, acquired from GE and Siemens MRI systems, with field strength of 1.5/3T.
Analysis was performed using Matlab R2017a and FMRIB Software Library (FSL).
Preprocessing: included intensity normalization and defining a bounding box around tumor area by setting six landmark points ~2 cm from the enhanced tumor area.
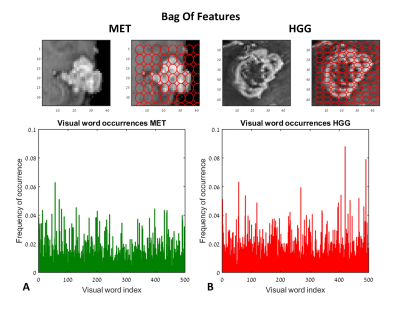
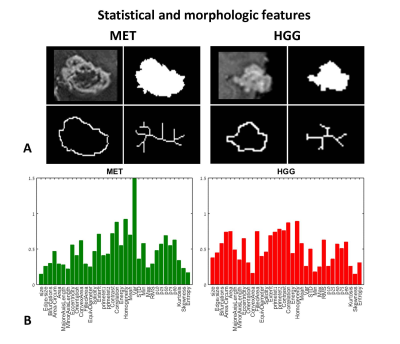
Features Extraction: Various features were extracted: 1. Bag-Of-Features7, with number of visual words = 500, grid point selection method, with grid step size (in pixels) = [5 5], extracted from the mid-slice of the defined bounding box and 2. Statistical-&-morphologic features including: gray level co-occurrence matrix (GLCM) features8, histogram based features, area, axis lengths, number of bifurcations, eccentricity, solidity etc. extracted from the enhanced tumor area, segmented from the mid-slice of the defined bounding box.
Classification: Feature selection was performed based on a statistical tests (Mann-Whitney U-test / t-test), and principal-component-analysis (PCA), and data standardization, classification was performed between HGG and MET and between breast and lung metastasis. Classification was performed using five machine ensemble classifiers. The input data for classification was given in 3 schemas: 1. BoF + statistical-&-morphologic features (n=535), 2. Bag-Of-Features (n=500), 3. statistical-&-morphologic features (n=35). Results were evaluated using a 5-fold cross-validation of the data. Sensitivity, specificity, accuracy and area under the receiver operating characteristic curve (AUROC) were calculated for each condition.
RESULTS
Classification of HGG and MET: Figure 1 and 2 demonstrate the BoF (Fig.1) and statistical-&-morphologic features (Fig.2) extracted from HGG and MET. Significant group differences were detected for 232/500 BoF and 27/35 statistical-&-morphologic features. Classification results are given in Table 1. The best classification results (marked in bold) were obtained using statistical-&-morphologic features and using Ensemble Subspace Discriminant classifier with a mean accuracy=83%, sensitivity=87%, specificity=81% and AUROC=0.92%.
Classification of breast and lung MET: Significant groups differences were detected for 60/500 BoF and 12/35 statistical-&-morphologic features. Classification results are given in Table 2. The best classification results (marked in bold) were obtained using BoF + statistical-&-morphologic features and using Ensemble Bagged Trees classifier with a mean accuracy=79%, sensitivity=76%, specificity=80% and AUROC=0.78%.
DISCUSSION & CONCLUSION
The proposed study suggests a non-invasive, cost effective clinical decision-support tool, for automatic classification of brain tumors. Results demonstrate the clinical potential of using automatic classification based on conventional imaging to classify brain tumors in newly diagnosed patients. BoF analysis may be superior over other methods for features extraction due to its minimal demands for user intervention, its robustness and computational efficiency. Similar methods may be further implemented to improve diagnosis of metastatic lesions of other cancers, as well as cancers originating in other body parts.Acknowledgements
To Vicki Myers for editorial assistanceReferences
- Giese, A. and M. Westphal, Treatment of malignant glioma: a problem beyond the margins of resection. Journal of cancer research and clinical oncology, 2001. 127(4): p. 217-225.
- O'NEILL, B.P., et al. Brain metastatic lesions. in Mayo Clinic Proceedings. 1994. Elsevier.
- Davis, F.G., B.J. McCarthy, and M.S. Berger, Centralized databases available for describing primary brain tumor incidence, survival, and treatment: Central Brain Tumor Registry of the United States; Surveillance, Epidemiology, and End Results; and National Cancer Data Base. Neuro-oncology, 1999. 1(3): p. 205-211.
- Surawicz, T.S., et al., Descriptive epidemiology of primary brain and CNS tumors: results from the Central Brain Tumor Registry of the United States, 1990-1994. Neuro-oncology, 1999. 1(1): p. 14-25.
- Posner JB. Management of brain metastases. Rev Neurol. 1992;148:47787.
- Patchell RA. et al. A randomized trial of surgery in the treatment of single metastases to the brain. N Engl J Med. 1990;322:494500.
- Csurka, G., et al. Visual categorization with bags of keypoints. in Workshop on statistical learning in computer vision, ECCV. 2004. Prague.
- Haralick, R.M. and K. Shanmugam, Textural features for image classification. IEEE Transactions on systems, man, and cybernetics, 1973(6): p. 610-621.
Figures