1515
Adult eye segmentation in MRI using active shape model: towards a personalized eye model for radiation treatment of uveal melanoma1Proton therapy Center, Paul Scherrer Institut (PSI), ETH Domain, Villigen, Switzerland, 2Ophthalmic Technology Laboratory, ARTORG Center of the University of Bern, Bern, Switzerland, 3Radiology Department, Centre d’Imagerie BioMédicale, Lausanne University Hospital, Lausanne, Switzerland, 4Signal Processing Laboratory (LTS5), Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Synopsis
We aim to construct a 3-dimensional patient-specific eye model from MRI data in order to later be integrated into proton radiation treatment planning. Our major challenge is the presence of motion, as subjects are awake and physiologically blink eyes. Additionally, fixing a point during acquisition might be challenging for some patients with ocular tumors. As such, in this study we evaluated an Active Shape Model (ASM) segmentation on a data set of 31 subjects, including 3 uveal melanoma (UM) patients. Quantitative evaluation in comparison with manual delineations shows good accuracy, even for images with the presence of UM and tantalum clips.
Introduction
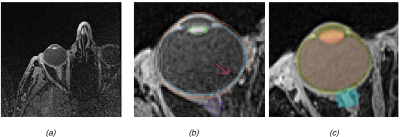
Over the past few decades radiotherapy has become used more widely in the conservative treatment of uveal melanoma. The highly localized dose distributions of proton radiation therapy (PT), provides high rates of local tumor control with acceptable ocular morbidity1. For the best planning processes, the localization and segmentation of eye structures must be applied to reduce inter-observer variability in boundary definitions, especially in posterior tumors with hemorrhage and tantalum clips. Practically, 3D MRI with high spatial resolutions provides a large amount of information about the eye anatomy and the tumor. Although, 3D MRI is not currently integrated in PT treatment plans, it is used for diagnosis and follow up due of its contributions to three dimensional representation of the tumor volumes and for the organ at risk delineation. In our previous work2, an Active Shape Model (ASM) was applied to pediatric eye retinoblastoma. Here, we are focus on the application of this image processing schema for awake adult eyes wherein motion and bias field artefacts in MRI acquisition or bigger eye volume (see Fig1(a, b)) can be observed.Materials and Method
Our data set is composed of 28 healthy eyes of volunteers aged 29±5.4 years old (range [23-46]) and 3 eyes from patients with UM (63, 63, 71 years old). All subjects gave written informed consent prior to participation. Cohort median eye size is 25.2 mm of diameter (range [23-26.5]). 1.5T MR T1-weighted VIBE images were acquired with a Siemens scanner using a surface loop coil and following acquisition parameters: TR/TE 6.55/2.39 ms; FA 120; 0.5x0.5x0.5 mm3; 80 slices. Manual delineations of lens, Vitreous Humor (VH), sclera and optic nerve head were done using the radiation therapy planning software Velocity TM (Varian Medical System). Image pre-processing pipeline consisted in cropping eye region, anisotropic diffusion denoising, bias field correction and histogram equalization. Model generation is done by combining all the surfaces extracted from manual delineation, generating an ASM that encodes shape and intensity. Segmentation of a subject is reduced to an optimization problem where the gradient between the ASM and the subject is minimized2 (Fig1c).Results
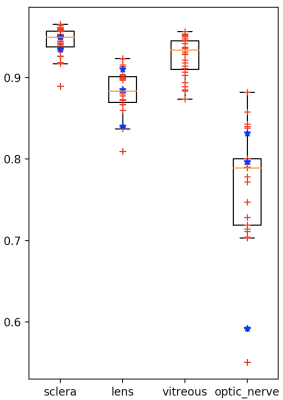
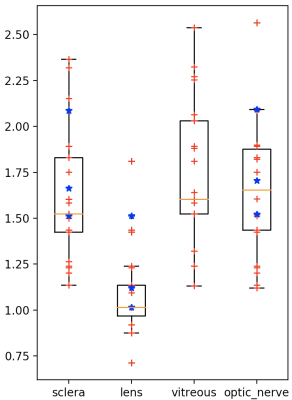
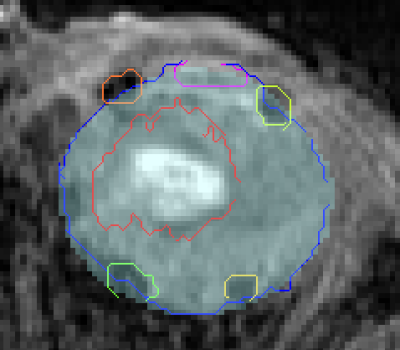
We perform a leave-one-out cross-validation (mixing both volunteers and patients). Boxplots of volume overlap (Dice similarity coefficient, DSC) and contour distance (Hausdorff distance, HD) were computed. Fig2 (respectively of Fig3) shows the result of DSC (of HD) measurement. Median of DSC (respectively of HD) were of 94.5% (1.75 mm) for the sclera, 92.6% (1.87 mm) for the vitreous, 88.2% (1.09 mm) for the lens and 77.1% (1.67 mm) for the head of optic nerve. Fig1(c) shows qualitative results on one volunteer and Fig4 on a patient with a large melanoma and 4 clips. The average computation time for an eye was of 10 seconds.Discussion and Conclusion
High volume overlap values were obtained for the segmentation of both the sclera and the vitreous humor, equivalent to previous reported results in children. The lens presents the best accuracy in terms of Hausdorff distance, while its slightly lower DICE are explained by its small size and from the fact that DICE number is sensitive to total volume. The results for optic nerve ROI delineation were less optimal. We decided to add this structure to our ASM even though available manual segmentations were not initially aimed for training but simply to approximately detect the head of the optic nerve. This region is also small and difficult as located nearby muscles. Some outliers can be identified in the lower bound, they are related the smallest eyes in the training set. Therefore, a larger dataset with a wider diversity in eye size and multimodal images could provide better segmentation results. Our works is the first attempt to automatically segment adult eyes including patients with UM. Our results show that automated ASM segmentation can succeed in presence of motion, tumors and tantalum clips. It allows for potential improvement in treatment planning and diagnosis by reducing the time spent by radiation oncologists. Our pipeline can be also extended to other ocular magnetic fields 3T or 7T by creating a corresponding training set. However, further developments are needed to for precisely segmenting the tumor and clips (eg. including other MR contrasts for tumor segmentation or a specific clip ASM model).Acknowledgements
This work is funded by the Swiss Cancer Research foundation (grant no. GAP-CRG-201602).References
1. Egger E., et al. Maximizing local tumor control and survival after proton beam radiotherapy of uveal melanoma. Int J Radiat Oncol Biol Phys 2001; 51(1):138-47.
2. Ciller C., et al. “Automatic Segmentation of the Eye in 3D Magnetic Resonance Imaging: A Novel Statistical Shape Model for Treatment Planning of Retinoblastoma”, Int J Radiat Oncol Biol Phys 2015; 92(4): 794–802.
Figures