1466
Impact of tissue image segmentation errors on SARAsha Singanamalli1, Matthew Tarasek1, Qin Liu2, Desmond Yeo1, and Thomas Foo1
1GE Global Research, Niskayuna, NY, United States, 2GE Healthcare, Waukesha, WI, United States
Synopsis
In this study, we evaluate the sensitivity of peak and global SAR to false positive (FP) and false negative (FN) errors in segmentation for three major brain tissue types: Gray Matter (GM), White Matter (WM) and Cerebrospinal Fluid (CSF). Voxel probability maps of GM, WM and CSF are thresholded at various intervals to generate multiple anatomical head models from a simulated T1w MRI dataset. FP and FN errors in segmentation are evaluated for each anatomical model with respect to the ground truth. Electromagnetic simulations are performed to relate these errors to peak and global SAR values at 3T.
Purpose
Radiofrequency
(RF) power deposition, quantified using Specific Absorption Rate (SAR), may pose
a safety concern in magnetic resonance imaging (MRI). The use of multiple generic
human body models (HBM) in EM simulations can yield estimates of global and
local SAR safety limits for transmit coils, but practical implementation
requires the SAR limits to be conservative. Recent advances[1] show a trend
towards subject-specific HBMs for more accurate SAR estimation, which is
particularly important for parallel transmit coils as the increased degrees of
freedom for RF transmit amplitudes and phases compound with uncertainty in the
HBM. While various registration and segmentation based approaches can be used
to generate subject-specific HBMs, each approach invariably introduces tissue
segmentation errors. In this work, we investigate downstream effects of such errors
on SAR prediction.Methods
Simulated T1w MRI, generated using anatomical models of normal brains, was acquired from the Brainweb database[2]. T1 data was simulated with the following parameters: SFLASH (spoiled FLASH) sequence with TR=22ms, TE=9.2ms, flip angle=30 deg and 1 mm isotropic voxel size. The corresponding discrete anatomical models provide a unique class label at each voxel that is most representative of that voxel. The model comprises of 11 class labels, including background, CSF, Gray Matter, White Matter, Fat, Muscle, Muscle/Skin, Skull, Vessels, Around Fat, Dura Matter, and Bone Marrow. Statistical Parametric Mapping (SPM)[3] was used to obtain voxel probability maps of Gray Matter (GM), White Matter (WM) and Cerebrospinal Fluid (CSF) from the simulated T1 volume. Six anatomical models, a few of which are shown in Figure 1, were generated by varying the probability thresholds for each tissue type. Segmentation errors for each of these tissues were evaluated with respect to ground truth using false positive error (FPE) and false negative error (FNE) metrics. A 16-rung high-pass birdcage head coil (dia. 37.0 cm, height 40.0 cm) in an RF shield was modelled using the FDTD software Sim4Life (ZMT, Zurich, Switzerland). The coil was tuned to 127.7 MHz with a loader phantom (σ=0.7 S/m, εs=78), and driven in quadrature mode in simulations using the ground truth anatomical model as well as the SPM output based models to evaluate differences in peak and global SAR.Results & Discussion
Results suggest that global SAR has stronger associations with segmentation errors than peak SAR. Plots in Figure 2 indicate that CSF FPE (R2 = 0.89) and FNE (R2 = 0.92) show the strongest linear association with global SAR, while GM and WM show moderate associations (0.4 < R2 <0.8). As shown in Figure 3, there was no strong relationship (R2 < 0.1) between peak SAR value and FPE or FNE. This is likely because peak SAR location varies with the different anatomical models and may therefore not be directly comparable. The slope of linear trendlines indicate that changes in CSF has the largest impact on global SAR, followed by GM and WM, the sequence of which is inversely associated with with tissue volume. In addition, the regression results show that FPE in CSF and WM is inversely correlated with global SAR whereas GM FPE is directly correlated with global SAR. On the contrary, FNE in CSF and WM is directly correlated with global SAR whereas GM FNE is inversely correlated with global SAR. Although the results from EM simulations provide some intuition for the impact of various types of segmentation errors on SAR estimates, it is important to further examine the impact on temperature changes in order to better understand the extent and type of allowable segmentation errors. Towards that end, thermal simulations are underway which will then allow us to connect segmentation errors with expected temperature change.Conclusion
In this work, we study the impact of over- and under-segmentation in CSF, WM and GM on SAR estimates using simulated T1w MRI. While there was no apparent linear relationship between peak SAR and segmentation errors, linear associations were found between the segmentation errors and global SAR. These results imply that patient-specific anatomical models can be systematically tailored to favor the types of errors that result in over-estimation of global SAR in order to ensure patient safety while moving towards subject specific anatomical models. However, additional work needs to be done to better understand the relationship between segmentation errors and peak SAR.Acknowledgements
No acknowledgement found.References
[1] Jin, J., Liu, F., Weber, E., & Crozier, S. (2012). Improving SAR estimations in MRI using subject-specific models. Physics in medicine and biology, 57(24), 8153. [2] Aubert-Broche, B., Griffin, M., Pike, G. B., Evans, A. C., & Collins, D. L. (2006). Twenty new digital brain phantoms for creation of validation image data bases. IEEE transactions on medical imaging, 25(11), 1410-1416. [3] Penny, W. D., Friston, K. J., Ashburner, J. T., Kiebel, S. J., & Nichols, T. E. (Eds.). (2011). Statistical parametric mapping: the analysis of functional brain images. Academic press.Figures
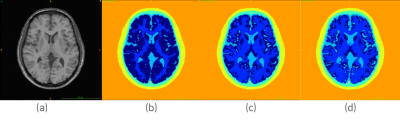
Axial view of (a) simulated T1w MRI and the corresponding (b-d) tissue
segmentations maps generated by thresholding tissue probability maps at various
values
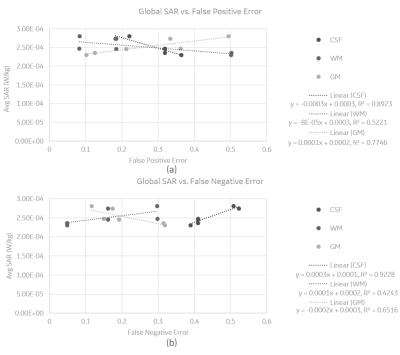
Global SAR vs. (a) False Positive and (b) False Negative Error in CSF,
WM and GM segmentation.
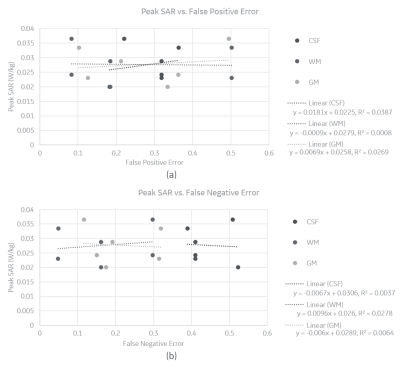
Peak SAR vs. (a)
False Positive and (b) False Negative Error in CSF, WM and GM segmentation.