1442
Performance of different classifiers in the diagnosis of benign and malignant bone tumors based on MR diffusion kurtosis imaging1Clincial Science, Philips Healthcare, Beijing, China, 2Radiology Department of First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Synopsis
Recently, the AI (Artificial Intelligence) is popular in the clinical diagnosis based on medical imaging. The major target is to identify or classify the disease condition through the features extracted from the clinical images. Different algorithms (or classifiers) can be applied to classify the disease and the performance might be different for a specific clinical issue. In this work, we tried to investigate the performance of different classifiers in the diagnosis of benign and malignant bone tumors based on MRI diffusion kurtosis imaging.
Introduction
Recently, the AI (Artificial Intelligence) is popular in the clinical diagnosis based on medical imaging. The major target is to identify or classify the disease condition through the features extracted from the clinical images. In the view of AI, the features extracted from the images are very necessary for the classification. Besides, different algorithms (or classifiers) can be applied to classify the disease and the performance might be different for a specific clinical issue. In the clinical studies, DKI (diffusion kurtosis imaging) and DKI-derived parameters has been proved to be stable and efficient biomarkers for tumors which were related to the cell density and tissue complexity. In this work, we tried to investigate the performance of different classifiers in the diagnosis of benign and malignant bone tumors based on the features extracted from MRI DKI-derived parameters.Methods
This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, Zhengzhou, China. Totally 42 patients (22 males and 20 females aged 33.2±18.6 years old) including 19 benign and 23 malignant bone tumor patients were recruited in this study. All the patients were diagnosed through MRI examination and finally verified by the tumor biopsy. The DKI imaging was performed with a 3T MR scanner (Ingenia, Philips Healthcare, Best, the Netherlands) with b values of 0,600,1200s/mm2 and 15 gradient directions. DKI post-processing was carried out by suing DKE software (Diffusion Kurtosis Estimator, Version 2.6.0). DTI-derived (MD,FA, AD,RD) and DKI-derived (MK,AK, RK) parameters were obtained. A 3D whole volume textures were extracted for the above parameter maps and diffusion images with B=0,600 and 1200 based on home-developed Matlab scripts. Forty-three features were obtained for each image. And totally 43×10 feature were acquired for the classification. Fisher score were firstly adopted for the features reduction and after that classifiers including SVM (Support Vector Machine) with linear, radial basis function (RBF) and sigmoid kernel, LDA (Least Discrimination Analysis), NB (Naïve Bayes) and RB (Random Forest) were adopted and compared for the diagnosis of benign and malignant bone tumors.Results
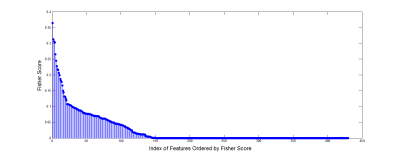
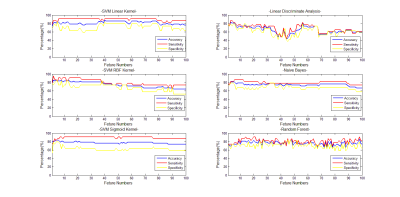
The fisher scores of the 43×10 features were shown in Figure 1. The classification results of different classifiers were shown in Figure 2.Discussion
Fisher score gives the importance of each feature and the features with big scores will give a relative good classification results. But for the features with small scores contribute little to the classification. Different classifiers showed different classification accuracy, sensitivity and specificity along the selected feature numbers. SVM with linear and RBF kernel, NB and RF presented a relative stable and good classification along the feature numbers. But the SVM with sigmoid kernel and LDA give a poor result. Besides, the appropriate feature numbers will give a balanced high accuracy, sensitivity and specificity. All in all, the textures extracted from DKI-derived parameters can give a good identification of benign and malignant bone tumors. And the results indicated that for this specific clinical issue, an appropriate classifier and feature numbers should be taken into consideration for a better classification results.Conclusion
For a specific clinical issue, different classifiers showed different performances. And an appropriate classifier and feature numbers would contribute to the classification results.Acknowledgements
No acknowledgement found.References
[1] Peter Raab, Elke Hattingen ,Kea Franz, et al. Cerebral Gliomas: Diffusional Kurtosis Imaging Analysis of Microstructural Differences[J].Radiology,2010,254(3):876-881.
[2] Vallières, M. et al. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Physics in Medicine and Biology, 2015, 60(14), 5471-5496. doi:10.1088/0031-9155/60/14/5471