1371
T2* Enhancement for Multi-Echo Data Image Combination -- Using least squares for echo prediction1Siemens Shenzhen Magnetic Resonance Ltd, Shen Zhen, China, 2Siemens Healthcare China Ltd, Shang Hai, China
Synopsis
This work provides a virtual echo prediction method for Multi Echo Data Image Combination (Medic) based on least square estimation. The strong dependences between multi-echoes in Medic sequences are used to predict virtual echoes with assumed echo times, and then such predictions are combined with real acquired echoes for heavier T2* contrast enhancement.
Purpose
Multiple Echo Data Image Combination (MEDIC) is a heavily T2* weighted spoiled gradient echo sequence, in which unipolar frequency encoding gradients are used to achieve flow compensation and off resonance effects, and multiple echoes acquired in one measurement are combined to improve the signal to noise ratio. So the multi-echo combination is potentially useful in imaging of cartilage in joints[1][2] Although the more echoes it combined, the heavier T2* contrast will be, yet echo numbers are usually less than 6 for scan time consideration. In this work, a linear prediction module based on least square estimation is used for extended echoes prediction considering the strong dependences between multi-echoes. In vivo experiments are performed to validate the hypothesis that the predicted echoes bring additional information and could be used for contrast enhancement.
Method
Conventionally, the relation between multi echoes in Medic is simply module as a T2* attenuation process, while in this study multi echoes formation process is treated as a time-series. The approach to predict future values of a time-series is based on linear prediction module [r.1], which can be intuitively understand by deriving from conventional T2* attenuation process attached with a time-invariant deviation (r.2 and r.3). If past echoes are available, then the problem of finding coefficients a1 and a2 can be solved using least squares. Finding C = [a1; a2] can be viewed as one of solving an overdetermined system of equations. We seek a second order linear predictor (high order predictor needs more acquired echoes), and then the overdetermined system of equations is given by [r.4]. Once the coefficients are found, echo (n) for n > N can be estimated using the recursive difference equation [r.1]. Since SNR drops very fast in predicted echoes, so an empirical filter is used to mitigated the disorders, then the predicted echoes are combined (using sum of square method)with real acquired ones to enhance T2* contrast.
$$$echo(n) = a_{1}*echo(n-1)+a_{2}*echo(n-2) \ \ \ \ \ \ \ \ \ \dashrightarrow (r.1)\\echo(n)=e^{\frac{-\triangle t}{T*_{2}}} *echo(n) + dev(\triangle t) \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \dashrightarrow\left(r.2\right)\\echo(n)=e^{\frac{-\triangle t}{T*_{2}}} *echo(n) + dev(\triangle t)\ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \dashrightarrow (r.3) \\\begin{vmatrix}y(3) \\y(4)\\.\\y(n)\end{vmatrix}= \begin{vmatrix} y(2) \ \ y(3)\\y(3)\ \ y(2)\\.\ \ .\\Y(n)\ \ y(n-1) \end{vmatrix}* \begin{vmatrix}a_{1} \\a_{2}\end{vmatrix}\ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \dashrightarrow (r.4)$$$
Result
In-vivo studies were performed on a 1.5 Tesla (T) MAGNETOM Amira MRI scanner (Siemens Healthineers) with volunteers admitted knee examinations. An 18-channel iTx Knee coil was used for data acquisition. Protocol parameters are listed below: FOV = 190x142mm, TR = 500ms, TE = 23ms, average = 2, BW/pixel = 230Hz, image slices = 5, phase oversampling = 10%, distance factor = 30%, voxel = 0.5x0.5x0.5mm, base resolution = 192, combined echoes = 5, scan time 1’53’’. The result showing [Fig2, b1-b5] indicated that signal in bones drops very fast in predicted echo series ,while signal in fluid, cartilage and muscle still keep a relative high level, contrast information between interest tissues still exist, which could be combined with real echoes to enhance the contrast between synovial fluid and articular cartilage[Fig3, e1 and e2].
Conclusion
The multi-echo formation process of Medic sequence was modeled as a linear-shot-time invariant system, and least square estimation method was used to predict more virtual echoes, providing more information and heavier T2* contrast. However there is still no indication that the enhanced image can completely take place of the conventional, and clinical evaluations may still be required to validate its value in practice.
Acknowledgements
The author would like to thank Dr. Chen Shi (HKU) for her helpful suggestions and comments on this topic.
References
[1]Bhawan K Paunipagar et,al. Imaging of articular cartilage. Indian J Radiol Imaging. 2014 Jul-Sep; 24(3): 237–248.[2]Marius R. Schmid et, al. Imaging of Patellar Cartilage with a 2D Multiple-Echo Data Image Combination Sequence, American Journal of Roentgenology,June 2005, Volume 184, Number 6
Figures
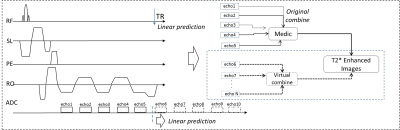
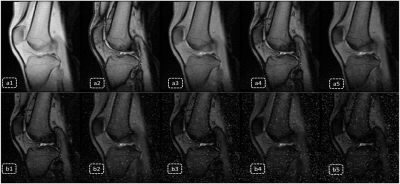
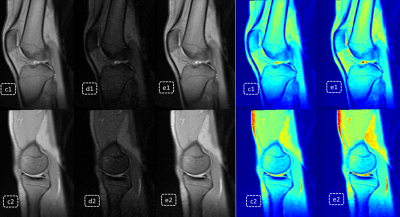
Fig3. Virtual enhancement result; c1 and c2 and product Medic images combined by 5 real echoes; d1 and d2 are images combined by 5 predicted echoes; e1 and e2 and virtual enhanced result with combination of both real echoes and predicted ones; it’s obvious both signal intensity and contrast between synovial fluid and articular cartilage are enhanced on e1 and e2.