1328
Highly Accelerated (R=14) Water Reference Acquisition for High Resolution 1H MRSI using Compressed Sensing1Max Planck Institute for Biological Cybernetics, Tuebingen, Germany, 2IMPRS for Cognitive and Systems Neuroscience, Eberhard-Karls University of Tuebingen, Tuebingen, Germany, 3Department of Physics, Ernst-Moritz-Arndt University Greifswald, Greifswald, Germany
Synopsis
In this study, the acquisition of a high resolution (64x64) water reference MRSI data is accelerated by a factor of R=14 using compressed sensing. The results show that this highly accelerated water reference can reliably be used for eddy current and phase correction purposes, as well as internal referencing and quantification. This enables the acquisition of the high resolution water reference MRSI data in 80 seconds at 9.4T.
Introduction
Proton magnetic resonance spectroscopic imaging (1H MRSI) is susceptible to time-dependent magnetic field distortions produced by eddy currents. To retrospectively correct for this, usually a non-water suppressed reference spectroscopic image is acquired and is afterwards used for eddy current and automatic phase correction. Additionally, for quantification purposes, the signal obtained from the unsuppressed water peak can be used for internal water referencing.
Despite the advantages of having a water reference spectroscopic image for every 1H MRSI dataset, this naturally comes at the price of a significantly prolonged scan time. However, due to the very sparse nature of water reference spectra, the acquisition can potentially be highly accelerated. Therefore, the aim of this work is to highly accelerate the acquisition of water reference images (with R=14) for application in the human brain at 9.4T through compressed sensing. This will enable the acquisition of a high resolution (64x64) water reference image in about 80 seconds.
Methods
Three healthy volunteers were scanned on a whole-body Siemens 9.4T human scanner with an in-house developed 16Tx/31Rx RF head coil [1]. Single-slice 1H MRSI data were acquired from each volunteer using a slice-selective 1H FID sequence [2,3,4] with no lipid or outer volume suppression. The acquisition parameters were: 64x64 matrix size; FOV = 200x200 mm; 10 mm slice thickness; TE= 1.5 ms, and TR= 300 ms. The acquisition time for the water-suppressed MRSI was approximately 15 mins.
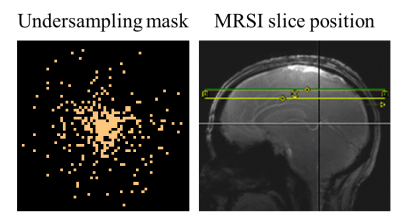
For each subject, two water references were acquired: one with reduced-resolution having 2x2 times lower spatial resolution than the MRSI dataset (as commonly done in the field), and a compressed sensing accelerated one at the full resolution (64x64) using a random variable-density undersampling mask (R=14) shown in Figure 1. The acquisition time for the water reference scans were 3.75 minutes for the reduced resolution, and 1.2 minutes for the compressed sensing approach.
For one of the subjects, a fully sampled water reference was also acquired with the same matrix size as the MRSI data and was used for comparison. The randomly undersampled k-space data was reconstructed using compressed sensing optimization [5] with a data consistency term, a total variation term and a wavelet sparsity term [6].
Results/Discussion
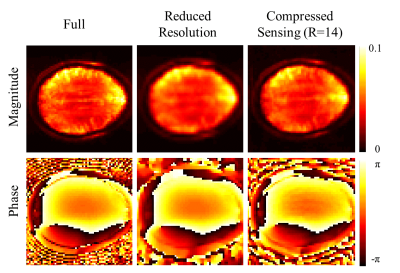
Figure 2 shows the magnitude and phase water reference images for the fully sampled data, the reduced resolution and the compressed sensing accelerated data. While the reduced resolution shows less anatomical details, the R=14 accelerated image preserves more anatomical details and resembles the fully sampled case better. All of the phase images are in good agreement to the fully sampled high resolution water image.
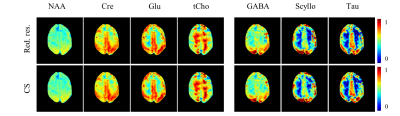
Each of the different water references were used for eddy current and phase correction of the MRSI data. The resulting metabolite maps for four major metabolites and three lower concentrated metabolites, namely, GABA, Scyllo-inositol and Taurine are shown in Figure 3. The difference between the reduced resolution and compressed sensing for eddy current and phase correction is virtually indistinguishable in the metabolite maps.
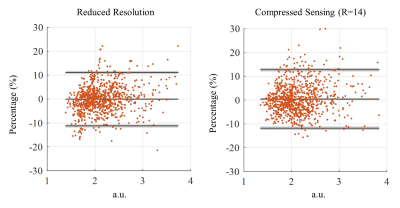
To quantitatively evaluate the change in the water peak amplitude resulting from compressed-sensing acceleration versus reduced resolution water reference acquisition, Bland-Altman plots comparing each of these methods to the fully sampled high resolution water reference spectroscopic image are shown in Figure 4. Despite the fact that the compressed sensing in this case has an acceleration factor that is more than 3 times faster than the reduced resolution one, the agreement limits of the reduced resolution and compressed sensing (R=14) are similar (less than 10% for most voxels).
Conclusion
In this study, the acquisition of the water reference image that is often needed for MRSI studies was accelerated through compressed sensing. An acceleration factor of up to R=14 was compared to a reduced resolution approach (equivalent to R=4). The results show that for quantification and referencing purposes an acceleration factor of R=14 results in similar accuracy as the reduced resolution approach while reducing the scan time by a factor of 3.5. In addition, for eddy current and phase correction purposes the proposed method can be reliably used without any loss in accuracy. This results in an acquisition time of 80 seconds for a high resolution matrix size (64x64) which can in turn greatly reduce the overhead scan time for high resolution metabolite mapping.Acknowledgements
No acknowledgement found.References
[1] Shajan et al, MRM 2014
[2] Henning et al, NMR in Biomed 2009
[3] Bogner et al, NMR in Biomed 2012
[4] Nassirpour et al, NeuroImage 2016
[5] Lustig et al, MRM 2007
[6] Chang et al, ESMRMB, 2017
Figures