1319
Novel methodology for processing, quality assessment, and artifact mitigation of raw 2D Correlation Spectroscopy data1The Charles Stark Draper Laboratory, Inc., Cambridge, MA, United States, 2Electrical and Computer Engineering, Boston University, Boston, MA, United States, 3Center for Clinical Spectroscopy, Brigham and Women's Hospital, Boston, MA, United States, 4Psychiatric Neuroimaging Laboratory, Brigham and Women's Hospital, Boston, MA, United States, 5US Army Institute of Environmental Medicine, Natick, MA, United States, 6Harvard Medical Institute, Boston, MA, United States
Synopsis
2D Correlation Spectroscopy (COSY) can be used to identify and study coupled resonances that cannot be observed or distinguished in 1D NMR spectra. However, resources and literature on best practices for processing raw 2D COSY data are limited. In this work, we describe a novel pipeline of signal processing algorithms and visualizations for quality assessment and artifact mitigation designed specifically for raw 2D COSY data, including detection of residual H2O and lipid contamination, correction for drift across averages, and peak location correction to enable more accurate comparisons of metabolites across subjects.
Introduction
2D Correlation Spectroscopy (COSY) can be used to identify and study coupled resonances that cannot be observed or distinguished in 1D NMR spectra1. However, the increased information it provides also increases the complexity of the methods required to extract high quality 2D spectra from raw scanner data, and the literature on best practices for signal processing and quality assessment in 2D COSY data is limited. To address these issues, we have developed a suite of Python-based signal processing algorithms and visualizations for quality assessment and artifact mitigation designed specifically for 2D COSY data. These methods include detection of residual H2O and lipid contamination, correction for drift across averages, and peak location correction to enable more accurate comparisons of metabolites across subjects.2D COSY pre-processing
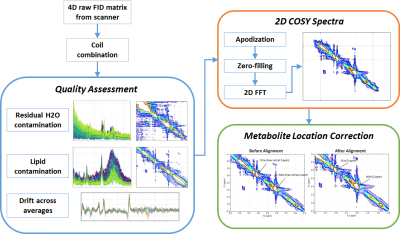
Figure 1 depicts the 2D COSY processing pipeline, which starts with raw scanner data containing the free induction decay (FID) signal acquired from each coil for each t1 increment, often with multiple repeated acquisitions (averages) for each of the increments. This creates a 4-dimensional complex matrix with dimensions [t1 increments x samples x averages x coils]. The dataset used in the following example was collected from a Siemens 3T scanner (TIM Skyra, VD11) using a 32 channel head coil. 2D COSY was acquired with a starting echo time of 30 ms followed by 64 t1 increments of 0.8 ms each, 8 averages per increment, and 2048 data points2. An SVD-based algorithm3 is used to derive coil weightings from the raw scanner data, which are applied to each average and t1 increment, creating a [64 x 2048 x 8] matrix.2D COSY signal quality assessment
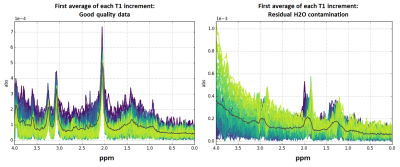
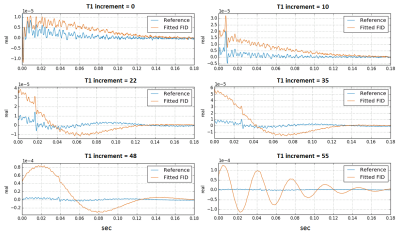
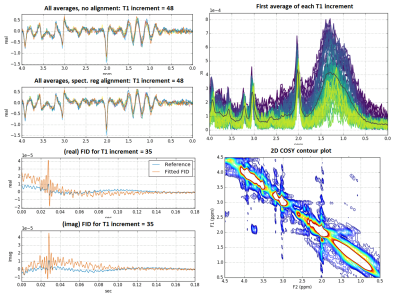
At this stage in the processing pipeline, failure of H2O suppression, lipid contamination, and issues with signal drift across averages can generally be detected visually, so we developed a series of simple diagnostic visualizations designed to rapidly highlight these kinds of issues. Contamination of the metabolite region of the spectrum by residual H2O changes the magnitude and shape of the signals in characteristic ways in the time and frequency domains4. It can begin at any point in the scan, thus affecting some increments and not others. Figures 2 and 3 depict diagnostic visualizations designed to highlight if and when issues with the H2O suppression process developed during the acquisition. These same views of the data can also be used to spot lipid contamination, as shown in Figure 4, and reveal issues with signal drift across averages. If drift is detected, it is fixed at this point using an implementation of the spectral registration algorithm5.Generating the 2D COSY spectra and metabolite location correction
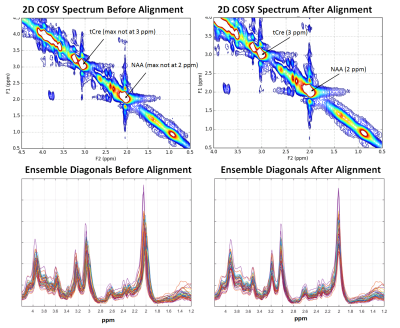
After coil weighting, quality assessment and correction for signal drift, we average the averages, apply apodization using a Tukey window, zero-fill the FIDs to increase the f1 frequency domain resolution, and perform a 2D Fast Fourier Transform. At this point, metabolite quantification can be performed; however, we’ve observed many cases where major metabolite peaks such as N-acetylaspartate (NAA) and Creatine are offset from their correct locations in the 2D parts-per-million (ppm) plane, as shown in Figure 5, top. These offsets are not necessarily the same for all individuals, making it difficult to compare metabolites across a population of subjects. To solve this problem, we developed a metabolite location correction algorithm designed to adjust peak locations, but not alter their amplitudes. We first extract the values that lay as close to the diagonal as possible in the 2D COSY matrix, and identify peak locations (in Hz) in the diagonal signal that are closest to a set of landmark singlets: N-acetylaspartate (2.008 ppm), Creatine (3.027 and 3.913 ppm), Choline (3.185 ppm), and myo-inositol (3.5217 ppm), for this dataset. In f1 and f2, we create a ppm axis piecewise between each pair of landmarks by finding the equation of the line intersecting the theoretical peak locations (in ppm) and the empirical peak locations (in Hz), creating a unique ppm mesh that maps the empirical peak locations to the theoretical ones. This mesh is then aligned to a common, uniformly sampled reference ppm mesh using 2D interpolation. Figure 5 (bottom) illustrates the effects of the processing pipeline for an ensemble of subjects.Conclusions
While there are toolboxes available for analysis and quantification of 2D COSY data, to our knowledge, there are few resources designed specifically for processing, quality assessment, and artifact mitigation of raw 2D COSY data. The pipeline described above provides a simple, semi-automated pre-processing approach we hope will enable more in-depth, accurate analysis of this information-rich data source.Acknowledgements
This study was funded by the Department of Defense Congressionally Directed Medical Research Program WX81-XWH-10-1-0835. The views expressed in this abstract are those of the authors and do not reflect the official policy of the Department of Army, Department of Defense, or the U.S. Government.References
1. Thomas, M.A., et al., Localized two‐dimensional shift correlated MR spectroscopy of human brain. Magnetic Resonance in Medicine, 2001. 46(1): p. 58-67.
2. Lin, A.P., et al., Changes in the neurochemistry of athletes with repetitive brain trauma: preliminary results using localized correlated spectroscopy. Alzheimer's research & therapy, 2015. 7(1): p. 13.
3. Rodgers, C.T. and M.D. Robson, Receive array magnetic resonance spectroscopy: whitened singular value decomposition (WSVD) gives optimal Bayesian solution. Magnetic Resonance in Medicine, 2010. 63(4): p. 881-891.
4. Kreis, R., Issues of spectral quality in clinical 1H‐magnetic resonance spectroscopy and a gallery of artifacts. NMR in Biomedicine, 2004. 17(6): p. 361-381.
5. Near, J., et al., Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magnetic Resonance in Medicine, 2015. 73(1): p. 44-50.
Figures