1308
Dictionary-Learning Compressed Sensing Reconstruction for an Anisotropic 3D Density-Adapted Radial Acquisition Sequence1Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 2Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 3Division of Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 4Erlangen Graduate School in Advanced Optical Technologies, Erlangen, Germany
Synopsis
Sodium magnetic resonance imaging requires dedicated acquisition techniques and reconstruction approaches due to the low in-vivo signal and ultra-short relaxation times. For this purpose a compressed sensing reconstruction technique using dictionary learning is applied to raw data acquired with an anisotropic 3D density-adapted radial acquisition sequence. The anisotropic acquisition allows an adjustment of projections in different directions to increase the in-plane resolution. In the following evaluation the possible benefits of the compressed sensing reconstruction using the increased in-plane resolution are shown for in-vivo sodium magnetic resonance imaging and quantification of 23Na.
Introduction
Sodium magnetic resonance imaging (23Na-MRI) requires dedicated acquisition techniques1 due to the low in-vivo signal and ultra-short relaxation times. Nevertheless, large voxel sizes are required to achieve sufficient signal-to-noise ratio (SNR). Acquisition techniques that enable anisotropic spatial resolution increase the perceivable in-plane resolution and have shown to be applicable for elongated structures like muscle or cartilage tissue2. Different kinds of compressed sensing4 reconstruction algorithms can be applied to improve 23Na-MR image quality by harnessing sparsity5,6. In this study, a 3D dictionary-learning compressed sensing reconstruction algorithm6 (3D-DLCS) was applied to images acquired with a 3D density-adapted acquisition sequence sampling a cuboid2 (DA-3DPR-C).Methods
23Na-MRI was conducted on a 3-T whole body system (Magnetom 3-T Skyra, Siemens Healthineers Erlangen, Germany). For the acquisitions and simulations an anisotropic DA-3DPR-C sequence was used, which allows an adjustment of the number of projections in x-, y- and z- direction and an increase of the in-plane resolution. Image reconstruction was performed using the iterative 3D-DLCS reconstruction (parameters: block-size: 43, dictionary size: 800, sparsity-term: 0.0025/50), which was adapted to use the anisotropic trajectory of the DA-3DPR-C sequence. Different undersampling factors were tested. Undersampling causes incoherent artifacts, which are corrected by the 3D-DLCS reconstruction.
Simulation Study
A simulated simplified calf-muscle phantom was created to evaluate the approach for quantitative 23Na-MRI. The simulated values are chosen similar to concentrations in muscle tissue (muscle: 20.0 mMol/l, muscle (increased concentration): 30.0 mMol/l, blood-vessels 50.0 mMol/l, boundary/fatty-tissue 10.0 mMol/l). A fully sampled data set with a nominal spatial resolution of 3x3x15mm3 was simulated for reference with the number of projections N = 11448 (52x52x20 voxels). White Gaussian noise was added to match the noise level of 23Na-MRI. For comparison 2.9-fold and 5.3-fold undersampled data sets are shown (c.f. Fig. 1). The median concentration and standard deviation is calculated for two regions of interest (ROI).
Phantom Study
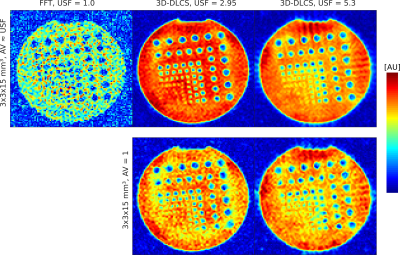
A resolution phantom filled with 0.3% NaCL solution was used to evaluate the image quality (c.f. Fig. 2). A fully sampled data set with a nominal spatial resolution of 3x3x15mm3 and N = 11448 (52x52x20 voxels) was acquired for reference (acquisition time (TA) = 9 min 32 s ). 2.9-fold and 5.3-fold undersampled data sets were acquired for comparison. The number of averages (AV) corresponds to the undersampling factor (USF) to ensure constant acquisition time and to compare image quality. Data sets without averaging were acquired and reconstructed with 3D-DLCS to evaluate the potential to decrease the acquisition time. Acquisition Parameters: TE/ TR = 0.30/50 ms; α = 80°; readout duration TRO = 10 ms.
In-vivo Study
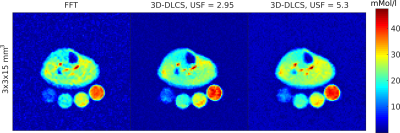
For an in-vivo evaluation of the imaging approach, 23Na-MR images of the right calf muscle were acquired. A fully sampled data set with N = 8264 (52x52x16 voxels) was acquired (TA: 7 Min 46 s) and a 2.9-fold and 5.3-fold undersampled data set with a resolution of 3x3x15mm3 were acquired matching the parameters of the phantom study (with AV ≈ USF).
Results and Discussion
The simulation study demonstrated that both reconstruction techniques yield concentration values close to the ground truth, whereas the 3D-DLCS algorithm resulted in a lower standard deviation of the evaluated ROIs (c.f. Tab.1). In the phantom and in-vivo study the 3D-DLCS reconstruction yields images with an improved image quality compared to the conventional FFT (c.f. Fig. 2 and Fig. 3). 3D-DLCS improves the distinction of the resolution rods in the phantom measurements (c.f. Fig. 2, AV ≈ USF). With the 2.9-fold and 5.3-fold undersampling of the phantom measurements TA has been reduced to TA = 3 min 14 s (for USF 2.9) and TA = 1 min 48 s (for USF 5.3) at the expense of a slightly decreased image quality (AV = 1) compared to 3D-DLCS reconstructions that include averaging (AV ≈ USF) (c.f. Fig. 2).
Conclusion
The 3D Dictionary-Learning-Compressed-Sensing reconstruction for an anisotropic 3D radial density-adapted acquisition sequence yields promising results to increase image quality and can simultaneously reduce acquisition time.Acknowledgements
No acknowledgement found.References
- Konstandin S, Nagel AM. Measurement techniques for magnetic resonance imaging of fast relaxing nuclei. Magn Reson Mater Phy 2014;27(1):5-19. 2.
- Nagel AM, Laun FB, et al. Sodium MRI using a density‐adapted 3D radial acquisition technique. Magn Reson Med. 2009; 62(6):1565-1573.
- Nagel, A. M. (2012): 3D Density-Adapted Projection Reconstruction 23Na-MRI with Anisotropic Resolution and Field-of-View. Proc. Intl. Soc. Mag. Reson. (2012): p. 674. 4.
- Lustig M, Donoho D, et al. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007; 58(6):1182-1195. 5.
- Gnahm C, Nagel AM. Anatomically weighted second-order total variation reconstruction of 23Na MRI using prior information from 1 H MRI. Neuroimage.2015; 105: 452-461. 6.
- Behl, N G R, Gnahm, C, et al. Three-dimensional dictionary-learning reconstruction of 23Na MRI data. Magn Reson Med. 2016; 75: 1605–16016.
- Wang Z, Bovik AC, et al. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13(4):600-612.
Figures
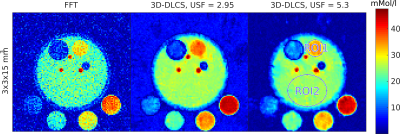
Figure 1: Images of a simplified muscle phantom simulation. The results are normalized to the simulated 23Na concentrations in the references and the median concentrations in ROI1 and ROI2 are compared.