1307
3D EPSI Hadamard spectral editing of GABA and GSH at 7T1Danish Research Centre for Magnetic Resonance, Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark, 2Center for Magnetic Resonance, Department of Electrical Engineering, Technical University of Denmark, Lyngby, Denmark
Synopsis
A 3D MRSI sequence was developed for simultaneous editing of GABA and GSH using a Hadamard editing scheme at 7T. 3D MRSI was performed using a 1D echo planar spectroscopic readout (EPSI). Volume selection was perfomed using a sLASER volume selection box using adiabatic refocusing pulses.
Introduction
Several highly interesting metabolites can be measured with increased accuracy at high field due to an increased sensitivity and spectral resolution (1). Due to technical limitations, only selected regions of interest are investigated, typically using single voxel spectroscopy (SVS). Due to the increases in SAR, B1/B0 inhomogeneity and physiological effects it has been difficult to perform spectroscopic imaging of larger regions. Several groups have also shown metabolic imaging with low-SAR sequences, such as pulse-acquire methods. However at such a short echo time, the measured spectrum is highly complicated by overlap between compounds.
Spectral editing sequences reduce complexity of the spectrum in order to increase accuracy of the measurements, at the cost of long TE and TR. MRSI with such methods can be made time efficient by using gradient readout trajectories (2). In this work we explore the use of echo planar spectroscopic imaging (3) for spectral editing, in order to increase specificity of metabolic measurements at 7T. For further acceleration, editing of multiple metabolites simultaneously was performed using a Hadamard encoded spectral editing scheme.
Materials and Methods
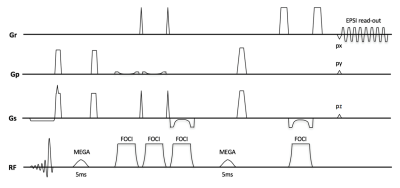
Experiments were performed on a 7T MRI system (Achieva, Philips Healthcare, Best, Netherlands) with a 32 channel receive head coil. All experiments were performed in accordance to the local ethical guidelines. A SVS MEGA-sLASER sequence with FOCI refocusing was combined with an echo-planar gradient waveform and two directional phase encoding.
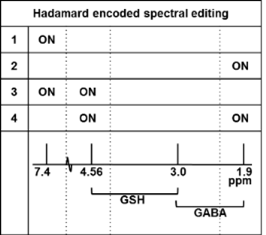
Single- and dual-band 180-degree sinc-Gaussian pulses (5ms duration, FW@95%=100Hz) were applied for refocusing of the J-coupling in GABA (1.9ppm) and GSH (4.56 ppm) in subsequent editing steps (5) (figure 1). The pulse duration was selected to have the B0 inhomogeneity range over the majority of the imaged volume covered within the FW@95% of the pulse. B1 sensitivity of the editing efficiency was simulated (VESPA, 6) and used in combination with a B1 map, as a scaling correction factor in post-processing. A 3D MRSI volume was placed transversal in the brain (fig 1) (FOV =240x165x120, matrix size=16x11x8, circular YZ k-space sampling, voxels size=15x15x15mm2, TR=3.5s, TE=74ms, editing cycles=4, total scan time = 17 min). The sLASER volume selection box was 110x75x80mm3 for lipid exclusion. The EPSI readout train (512x) was performed with spatial/spectral bandwidth of 64kHz/2666Hz, where both odd and even echoes were used after phase correction.
The protocol consisted of one water suppressed (VAPOR) scan and a water-reference scan (duration=3min) at the same TE, with the editing pulses turned off. Spatially localized spectra were reconstructed with Matlab software (Matlab, The MathWorks, Inc.) developed in-house with a Cartesian reconstruction in the spatial and temporal dimensions and including weak apodization, automatic phase and amplitude corrections over coil elements and spatial locations, and eddy current compensation. The four averages were phase and frequency aligned on a voxel-by-voxel basis.
Results and Discussion
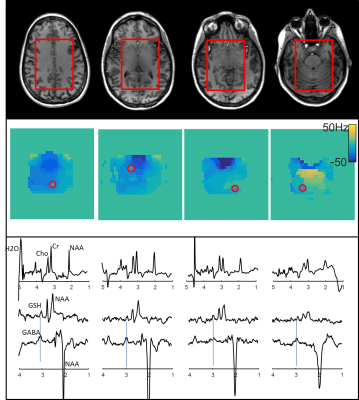
The residual B0-variation over the volume was in the range of +/-50Hz over the majority of voxels in the sLASER selection volume, although around 1% of the voxels, mainly in the frontal part in the lowest slices (close to the nasal cavities) exceeded this value, and were masked out of the analysis. Only negligible aliased peak ghosting from the Cartesian reconstruction was observed over the volume. Both GABA and GSH spectra could be reconstructed from the 4-step editing scheme. Figure 2 shows representative spectra from within the sLASER selection box.Conclusion
A MEGA-sLASER sequence was combined with an optimized echo-planar read-out for accelerated 3D MRSI mapping of both GABA and GSH while utilizing the high sensitivity at 7T field-strength. The results show in vivo mapping of the GABA inhibitory neurotransmitter and GSH dominant antioxidant. The technique may ultimately benefit studies of neurological and psychiatric disorders as well as studies of the healthy brain.Acknowledgements
This research is supported by the Danish Council for Independent Research [grant no. 6111-00349A].References
1. Stephenson M, Gunner F, Napolitano A, Greenhaff PL, MacDonald IA, Saeed N, Vennart W, Francis ST, Morris P. Applications of multi-nuclear magnetic resonance spectroscopy at 7T. World J Radiol 2011; 3(4): 105-113.
2. Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med 1995;33:34-40.
3. Magnusson P. ISMRM 2017:1255
4. Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DW J. Efficient Spectral Editing at 7T: GABA Detection with MEGA-sLASER. Magn Reson Med 2012; 68:1018–1025.
5. Chan KL, Puts NA, Schär M, Barker PB, Edden RA. Magn Reson Med. 2016 Jul; 76(1): 11–19.
6. https://scion.duhs.duke.edu/vespa/
Figures