1281
Comparison of adiabatic and non-adiabatic inversion pulses for lipid suppression in human calf muscle1Medical Physics Group, Institute of Diagnostic and Interventional Radiology, Jena University Hospital - Friedrich Schiller University Jena, Jena, Germany, 2Michael Stifel Center for Data-driven and Simulation Science Jena, Friedrich Schiller University Jena, Jena, Germany, 3Abbe School of Photonics, Friedrich Schiller University Jena, Jena, Germany, 4Center of Medical Optics and Photonics, Friedrich Schiller University Jena, Jena, Germany
Synopsis
Overlapping signal contributions originating from different metabolites with similar molecular structure is a common problem of in vivo 1H-MR spectroscopy with magnetic field strengths of ≤ 3 T. One prominent example is the “contamination” of the resonances of lactate with fat signals in 1H-MR muscle spectra. The goal of this work was to implement a MRS sequence with inversion recovery based adiabatic/ nonadiabatic lipid suppression and to test this approach in vivo in two different human calf muscles.
Introduction
Overlapping signal contributions originating from different metabolites with similar molecular structure is a common problem of in vivo 1H-MR spectroscopy with low and moderate field strength scanners (≤ 3 T). Taking advantage of the different longitudinal relaxation properties of overlapping metabolites such as, for instance, lipids and lactate in muscle, selective suppression can be achieved by using an adiabatic lipid nulling approach1. The current work demonstrates an implementation of this approach on a clinical whole-body MR scanner and compares the efficiency of lipid nulling in two human calf muscles with both adiabatic and non-adiabatic inversion pulses.Material and Methods
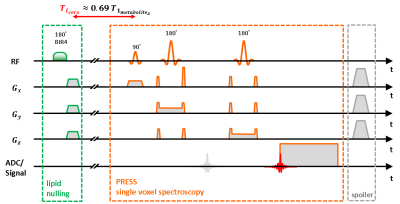
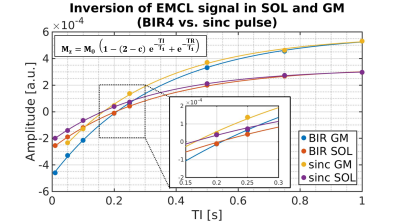
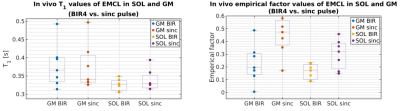
For implementation a conventional PRESS 1H-MRS sequence (Siemens IDEA, VE11-B) was used with a preceding spatially non-selective lipid nulling block based on the work of Hövener et al.1 (Fig. 1). Two different inversion pulses, an adiabatic BIR4 pulse2 and a non-adiabatic sinc-pulse, were implemented in order to evaluate the inversion efficiency and nulling of the EMCL lipid signal at 1.4 ppm in the presence of inhomogeneous B1 field distributions. In vivo measurements were conducted in seven male subjects (24-57 yrs.) with a clinical whole-body 3 T MR scanner (Prisma Fit VE11B, Siemens Healthineers AG) by using a flexible, double-tuned surface coil (1H/ 31P, ∅:11 cm, RAPID-Biomedical). Series of spectra were acquired with TR/TE = 5000/145 ms and varying inversion times (TI: 10 to 1500 ms) in the right M. Gastrocnemius Medialis (GM) and M. Soleus (SOL) (see Fig. 2 for voxel positions). After quantifying the spectra with the jMRUI 5.2 package (http://www.jmrui.eu), the T1 relaxation time constants and inversion efficiencies of the EMCL intensities were determined for both muscles and for each inversion pulse type by fitting the corresponding inversion recovery evolution with a mono-exponential model (Fig. 3). In the fit model, the inversion efficiency was expressed by means of the empirical factor c.
Results
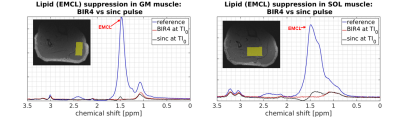
Fig. 3 demonstrates representative EMCL intensity evolutions (signal-vs.-TI-curves) for both adiabatic BIR4 and non-adiabatic sinc-shaped inversion pulses. Compared to sinc-pulse inversion, the TI-curves acquired with the BIR4 inversion pulses show different zero-crossings (TI0) for both muscles. Since the estimated EMCL T1 relaxation time constants in SOL and GM do not differ substantially between both inversion pulse types (see Fig. 4, left), the shorter TI0 with sinc-shaped pulse inversion can be ascribed to the lower inversion efficiency of the sinc-shaped pulse compared to the BIR4 pulse (see the apparently higher values of empirical factor c for sinc-shaped pulses in the right chart of Fig. 4). As seen in Fig. 2 appropriate EMCL suppression was achieved by using the adiabatic inversion pulses at TI0. In contrast, the sinc-pulse based inversion was associated with significant EMCL residuals in both GM and SOL spectra. This indicates the robustness of adiabatic pulses against B1 field inhomogeneities.Discussion and Conclusion
In this work, we demonstrated an improved suppression of EMCL resonances in the human calf muscle by using adiabatic BIR4 inversion pulses compared to non-adiabatic sinc-shaped pulses. Besides revealing better inversion efficiency, the BIR4 pulses also appear to be more robust against B1- inhomogeneities, which affect the adjustment of appropriate flip angles, specifically in rf surface coils. However, while being very effective in suppressing one selected lipid resonance with its characteristic T1 relaxation time, single pulse lipid nulling fails in fully suppressing multiple spectroscopic compounds, due to their different T1 relaxation times. Therefore, our next step will include the combination of adiabatic lipid nulling with spectral editing techniques, such as, e.g., the MEGA-PRESS sequence allowing additional broad-range lipid suppression in muscle spectra in vivo 2,3.Acknowledgements
Alexander Gussew acknowledges funding from the German Research Foundation (DFG, GU 1108/3-1).
Andreas Masek is supported by a graduate scholarship from the Friedrich‐Schiller‐University Jena (Landesgraduiertenstipendium).
References
1 Hövener J.B. et al.. Whole-Brain N-Acetylaspartate MR Spectroscopic Quantification: Performance Comparison of Metabolite versus Lipid Nulling. AJNR Am J Neuroradiol 2008; 29:1441-45.
2 Garwood M., Ke Y., Symmetric Pulses to Induce Arbitrary Flip Angles with Compensation for RF Inhomogeneity and Resonance Offsets. Journal of Magnetic Resonance 94 1991, 511-525.
3 Tschiesche K. et al.. Multimodal determination of load induced changes in the muscle - A combination of 1H MEGA-PRESS MRS and blood sampling. Proceedings of the International Society for Magnetic Resonance in Medicine, vol. 23, 2015.
Figures