1272
Standardized Parameterization of Echo-Planar Compressed Sensing MRSI Acquisition and Reconstruction1Department of Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States
Synopsis
Advanced MRSI acquisition strategies can be complex to implement and require customized reconstruction software, typically designed for a specific raw file format and that relies upon a priori knowledge of the specific implementation of the pulse sequence being applied. The ISMRMRD format1 has begun to address standardization in describing data acquisition parameters for different types of imaging data, but further development is needed. Here we build on this strategy by demonstrating XML encoding of parameters that describe flyback echo-planar, compressed sensing MRSI acquisitions being implemented on scanners from multiple vendors at UCSF that can be supported with generalized reconstruction software.
Purpose
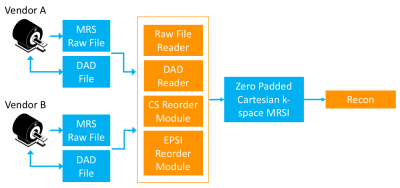
The purpose of this work is to define a set of standardized parameters that describe echo-planar, compressed sensing MRSI k-space trajectories2-3 and can be used for cross-platform implementation and reconstruction. In order to acquire and reconstruct data with randomly sampled k-space trajectories it is necessary for both the pulse sequences and reconstruction software to know the specifics of the trajectory and data ordering. At the current time this is addressed on a sequence specific basis for spectroscopic acquisitions and the software used to interpret the raw data or file formats for recording the parameters has to be tailored to individual uses. This can be resolved by defining a flexible set of parameters to describe a randomly sampled echo-planar acquisition for compressed sensing reconstruction that can be implemented on multiple vendor platforms and developing standardized software modules that are capable of reordering such acquisitions onto a zero-padded Cartesian kx,ky,kz,t grid suitable for reconstruction (Figure 1).Implementation
The k-space trajectory of the flyback echo-planar compressed sensing MRSI acquisition was defined by 3 parameter groups that describe 1) the 2D phase encoding trajectory, 2) the flyback waveform parameters for encoding the 3rd spatial and spectroscopic dimensions and 3) the blipped phase encoding pattern that defines the specific kx,ky points sampled along the flyback trajectories. This information was encoded in an XML Data Acquisition Descriptor (DAD) file4 with specific XML elements for each of these 3 parameter groups. Software libraries for reading and writing DAD XML files as well as data ordering modules used in reconstruction pipelines were implemented in C++ using the open-source SIVIC5 software package. Separate modules were developed for interpreting the blipped compressed sensing phase encoding scheme and for interpreting the flyback encoding (Figure 1). This enables the latter module to also be utilized for fully sampled EPSI acquisitions.
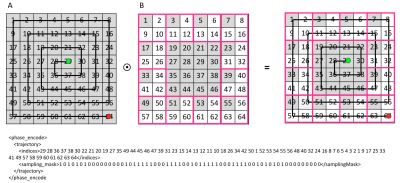
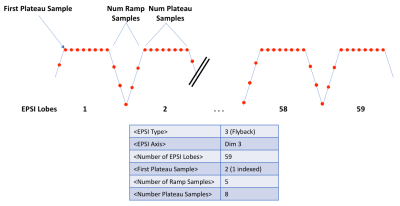
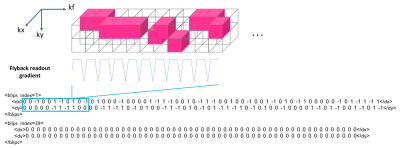
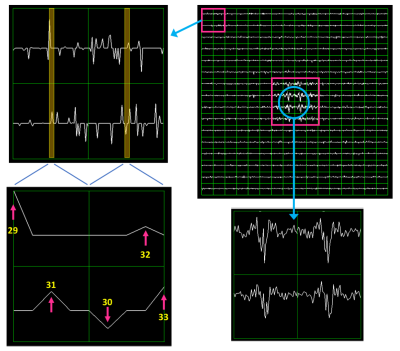
2D phase encoding is defined by a trajectory and a sampling mask element in the DAD file (Figure 2). The trajectory is defined by the linear indices corresponding to specific kx,ky pairs for each point in the trajectory, here shown for centric encoding. The sampling mask indicates which k-space indices are actually sampled. The EPSI waveform is defined by 6 parameters (Figure 3). Each EPSI waveform follows either a fully sampled trajectory along kx,ky,kz or a randomly undersampled path as prescribed by the DAD file <blip> elements (Figure 4) which define the blipped phase encoding gradient changes for each lobe along the flyback waveform. For example the EPSI readout starting at index 29 (Figure 2) fully samples the trajectory kx,ky,{kz}, whereas the readout starting at index 1 meanders randomly through a 2x2 block of kx,ky for each lobe of the waveform as indicated by the gradient blips for <blip index=1>.
Results
Software modules for reading and reordering 13C echo-planar compressed sensing data2 were implemented on a 14T Varian (Agilent) scanner in SIVIC. This initially used hardcoded trajectories and blip patterns to test the implementation. The reconstruction software was then refactored and generalized to read the parameters dynamically from a DAD file. This simplified the software modules and enabled them to be generalized to independently support arbitrary random sampling and echo-planar encoded acquisitions. The reordered, zero-padded data are shown in Figure 5. The EPSI reordering module, together with DAD file EPSI descriptors was able to support interpretation of fully sampled EPSI acquisitions from a GE scanner with DAD writing libraries linked into the custom PSD.Discussion
Raw data formats do not currently include sufficient information to reconstruct MRSI data that are acquired with advanced sampling strategies. Standardized parameterization of MRSI acquisition strategies such as parallel imaging, compressed sensing, and EPSI will streamline translation of sequences between vendors and enable utilization of these strategies using standardized software modules. Next steps in this work will include modifying echo-planar compressed sensing sequences, including for dynamic acquisitions, on GE and Bruker scanners to write DAD files describing these acquisition parameters and validating the reordering of raw data to a Cartesian k-space grid using the software modules used for the Varian implementation. Once tested on multiple scanner platforms, these descriptors will be proposed as additions to the ISMRMRD1 working group.Conclusion
Standardization of parameters required to describe common classes of complex MRSI sampling strategies will permit the development of software packages capable of interpreting the results and streamline development and translation of MRSI methods. These parameters are currently being encoded via a DAD/XML file, but will be proposed for incorporation into broader standards such as the ISMRMRD or DICOM. This will be critical for streamlining MRSI development and practical implementation.Acknowledgements
This work was supported by NIH grant P41 EB013598.References
1. Inati SJ, Naegele JD, Zwart NR, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn. Reson. Med. 2016;0(August 2015):n/a-n/a. Available at: http://doi.wiley.com/10.1002/mrm.26089.
2. Hu S, Lustig M, Balakrishnan A, et al. 3D compressed sensing for highly accelerated hyperpolarized (13)C MRSI with in vivo applications to transgenic mouse models of cancer. Magn. Reson. Med. 2010;63(2):312–21. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2829256&tool=pmcentrez&rendertype=abstract. Accessed August 25, 2014.
3. Larson PEZ, Hu S, Lustig M, et al. Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies. Magn. Reson. Med. 2011;65(3):610–619.
4. Olson MP, Crane JC, Larson P, et al. A Vendor-Agnostic MRSI Acquisition and Reconstruction XML Descriptor Format. In: ISMRM 25th annual meeting.; 2017. Available at: http://dev.ismrm.org/2017/5533.html.
5. Crane JC, Olson MP, Nelson SJ. SIVIC: Open-Source, Standards-Based Software for DICOM MR Spectroscopy Workflows. Int. J. Biomed. Imaging. 2013;2013:169526. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3732592&tool=pmcentrez&rendertype=abstract.
Figures