1198
Through-time and hybrid radial GRAPPA to improve the visualisation of velic motion in real-time speech MRI1Clinical Physics, Barts Health NHS Trust, London, United Kingdom, 2William Harvey Research Institute, Queen Mary University of London, London, United Kingdom
Synopsis
Aim: to investigate if radial GRAPPA could be used to: (a) improve velic motion visualisation when compared with standard protocols and reconstructions; (b) enable multislice imaging by sufficiently accelerating real-time MRI data acquisition.
Methods: datasets of healthy adult volunteers were acquired at 3T and reconstructed using through-time and hybrid GRAPPA methods.
Results: velic motion visualisation was superior in GRAPPA images than in images acquired using standard protocols and reconstructions. Multislice imaging (two slices) at 8fps per slice was achieved.
Conclusions: radial GRAPPA shows promise as a method for use in clinical imaging of speech.
Introduction
Clinical assessment of speech often involves imaging to visualise the motion of the velum. In the UK, videofluoroscopy and nasendoscopy are most commonly used, however studies have shown that velic motion can be visualised using real-time MRI (rtMRI).1,2
Multiview visualisation of the velum in speech is desirable as it can provide clinically relevant information about the symmetry of velic closure. Multislice rtMRI at temporal resolutions sufficient (>6.67fps)2 to accurately capture velic motion requires accelerated data acquisition. Accelerating rtMRI data acquisition while maintaining image quality is challenging, but has been achieved by using non-Cartesian trajectories and parallel imaging methods such as GRAPPA. 3-5
This study aims to investigate if radial GRAPPA could: (a) improve velic motion visualisation when compared with standard protocols and reconstructions; (b) enable multislice imaging by sufficiently accelerating rtMRI data acquisition.
Methods
Data acquisition: rtMRI datasets of four volunteers were acquired using a 3TX Achieva MRI scanner and a 16-channel neurovascular coil (Philips, Best, the Netherlands). Volunteers were scanned in a supine position while they counted from one to ten, said /za-na-za/, /zu-nu-zu/, /zi-ni-zi/, /a/ and /i/. Datasets undersampled by a factor (R) of 4, 5 and 10 (corresponding to temporal resolutions of 9, 11 and 22fps) and 200 frames of calibration data (R=1) were acquired using a single-slice spoiled gradient-echo sequence with a radial trajectory; FOV: 190×190mm2, in-plane pixel size: 1.9×1.9mm2, slice thickness: 10mm, TR/TE: 4.5ms/2.1ms, α: 10°.5 In addition, R=5 and calibration datasets of a volunteer were acquired using a multislice spoiled gradient-echo sequence with a radial trajectory; FOV: 192×192mm2, in-plane pixel size: 2.4×2.4mm2, 2 parallel slices, slice gap: 16mm, TR/TE: 7.7ms/1.74ms, temporal resolution: 8fps per slice.
Data reconstruction: All data were reconstructed using a radial GRAPPA method 3 implemented in MATLAB R2014b (MathWorks, NA). Initially, through-time GRAPPA (tt-GRAPPA) images were reconstructed. To investigate if the number of calibration frames could be reduced, hybrid GRAPPA (combined through-k-space and tt-GRAPPA) images were also reconstructed.
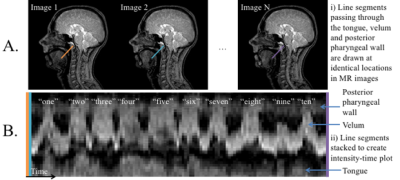
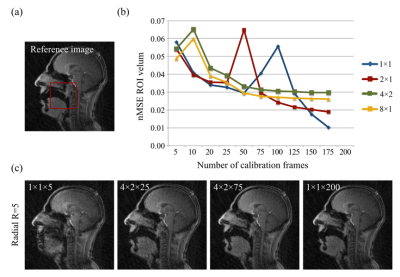
Data analysis: GRAPPA image quality was visually assessed and was considered to be adequate if the velum could easily be seen. Velic motion visualised in the images were represented using intensity-time plots (Figure 1). The effects of the following variables on image quality were investigated by comparing the normalised mean squared error (nMSE) in a region of interest containing the velum: the number of k-space segments and the number of calibration frames used in the GRAPPA method. GRAPPA images were compared with images acquired using standard protocols and reconstructions with similar spatial and temporal resolutions.
Results
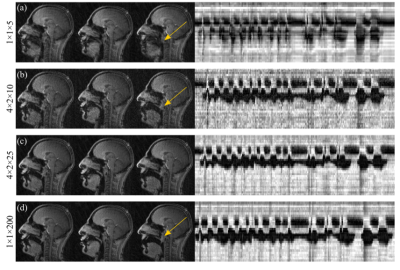
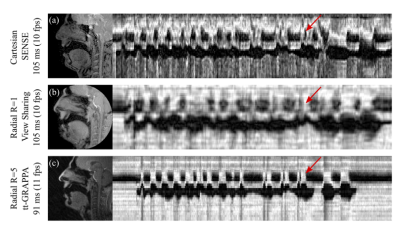
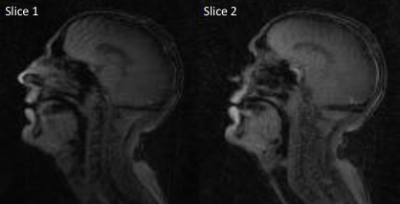
Radial GRAPPA methods were successfully applied to speech rtMRI data. Image quality was considered to be adequate in the R=4 and 5 tt-GRAPPA images, but not in the R=10 images. The image quality to temporal resolution trade-off was considered to be optimal in the R=5 images, therefore these images were used as the reference images in the nMSE calculations. nMSE increased as the number of calibration frames used in the GRAPPA method was decreased (Figure 2). Examples of different GRAPPA images, and the velic motion visualised in them are shown in Figure 3. Velic motion visualisation in R=5 tt-GRAPPA images was considered to be superior to that in images acquired using standard protocols and reconstructions (Figure 4). Multislice imaging of two parallel sagittal slices at a temporal resolution of 8fps per slice was achieved (Figure 5), as was multislice imaging of a midsagittal slice and an oblique slice.Discussion
The image quality to temporal resolution trade-off was considered to be optimal in the R=5 tt-GRAPPA images, however, acquiring 200 frames of calibration data was found to be time consuming (~100s) and could be challenging in patient populations who have difficulty repeating speech tasks for long periods of time. Using a hybrid GRAPPA method enabled the number of calibration frames to be reduced from 200 to 75 while maintaining adequate image quality (Figure 2), thus more than halved the calibration data acquisition time. The method would therefore be more suitable for use in clinical practice. Multislice imaging at a temporal resolution of 8fps per slice was achieved. This temporal resolution is greater than the minimum recommended for velic motion imaging.2 In all cases, image reconstruction was achieved in a time frame compatible with that of a clinical assessment (<360s).Conclusion
Radial GRAPPA methods were successfully applied to speech rtMRI data. The methods enabled improved visualisation of velic motion when compared to standard protocols and reconstructions, and accelerated data acquisition sufficiently to enable multislice imaging at 8fps per slice. In addition, image reconstruction was always achieved in a time frame compatible with that of a clinical assessment. Radial GRAPPA therefore shows promise as a method for use in clinical imaging of speech.Acknowledgements
Thank you to Matthew Clemence (Philips) for his technical support, and CLEFT and Barts Charity for investigator funding.
References
1. Scott A, Wylezinska M, Birch M, Miquel M. Speech MRI: Morphology and function. Phys Medica. 2014;30(6)604–618, 2014.
2. Lingala S, Sutton B, Miquel M, Nayak K. Recommendations for real-time speech MRI. J Magn Reson Imaging. 2016;43(1)28–44.
3. Seiberlich N, Ehses P, Duerk J, Gilkeson R, Griswold M, Improved radial GRAPPA calibration for real-time free-breathing cardiac imaging. Magn Reson Med. 2011;65(2)492–505.
4. Lingala S, Zhu Y, Lim Y, et al. Feasibility of through-time spiral generalized autocalibrating partial parallel acquisition for low latency accelerated real-time MRI of speech. Magn Reson Med. 2017; DOI: 10.1002/mrm.26611.
5. Freitas A, Ruthven M, Boubertakh R, Miquel M. Improved real-time MRI to visualise velopharyngeal motion during speech using accelerated radial through-time GRAPPA. MAGMA. 2017;30(1)17-18.
Figures