1197
Highly Accelerated Multi-Contrast 3D Isotropic MRI in 5 Minutes: A Feasibility Study for Multiple Sclerosis1Chinese PLA General Hospital, Beijing, China, 2Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Republic of Korea, 3Department of Brain and Cognitive Engineering, Korea University, Seoul, Republic of Korea, 4United Imaging Healthcare(UIH), Shanghai, China
Synopsis
Multiple sclerosis(MS) is a chronic disease that damages the nerves in the brain and results in multiple areas of scar tissues within the central nervous system. Multi-contrast structural MRI, which includes T1, T2, and FLAIR, has been routinely used in detecting MS lesions. Nevertheless, it is still challenging to accurately detect small MS lesions diffused over the entire brain due to the limitation of spatial resolution and long imaging time. The purpose of this work is to investigate the feasibility of achieving highly accelerated, multi-contrast 3D isotropic (~ 1.0 mm3) MRI (T1, T2, and FLAIR), which exploits sharable information across images, for detection of MS lesions over the whole brain roughly in 5 minutes.
Introduction
Multiple sclerosis(MS) is a chronic disease that damages the nerves in the brain and results in multiple areas of scar tissues within the central nervous system [1]. Multi-contrast structural MRI, which includes T1, T2, and FLAIR, has been routinely used in detecting MS lesions. Nevertheless, it is still challenging to accurately detect small MS lesions diffused over the entire brain due to the limitation of spatial resolution and long imaging time. The purpose of this work is to investigate the feasibility of achieving highly accelerated, multi-contrast 3D isotropic (~ 1.0 mm3 ) MRI (T1, T2, and FLAIR), which exploits sharable information across images, for detection of MS lesions over the whole brain roughly in 5 minutes.Methods
Data acquisition: MRI exams were performed in 5 patients using T1-, T2-, and FLAIR-weighted, whole brain imaging protocols on a 3.0 T System (uMR 770, UIH, Shanghai, China). Each data was acquired using an elliptical, incoherent variable density under-sampling pattern with a reduction factor (R) of 6. Imaging parameters common to all data were: FOV = 256x220x176mm3, encoding matrix = 256x220x176, and the number of receiver channels = 32. Those specific for T1-weighted imaging were: TR/TE/TI= 7.2/3.1/750ms, bandwidth=250Hz/pix; Those for T2-weighted imaging were: TR/TE=2000/330ms, TEeff=142ms, bandwidth=600Hz/pix, ETL=160, ESP=4.18ms; and those for FLAIR imaging were: TR/TE/TI =5000/461/1557ms, TEeff=154ms, bandwidth= 750Hz/pix, ETL =240, and ESP =3.72.
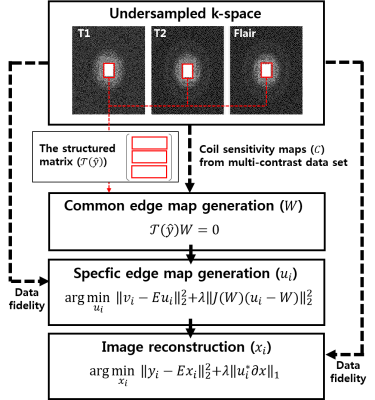
Image reconstruction: Multi-contrast MR image reconstruction with spatially adaptive regularization was performed from incomplete measurements (R = 6) by exploiting sharable information over the contrast dimension: edge structures and coil sensitivity information. Image reconstruction is composed of the following three steps: 1) generation of an edge map and coil sensitivity maps common to all contrasts, 2) generation of contrast-specific edge maps, and 3) image estimation using spatially adaptive regularization with the pre-defined, contrast-specific edge and coil sensitivity maps. The common edge map (W) in the step 1) was constructed by stacking row-vectorized patches in k-space over all contrasts, finding null space vectors, and combining null space images. It exploits the fact that the Fourier coefficients of the partial derivatives of an image satisfy a linear annihilating filter relation [2]. Coil sensitivity maps were generated from the full-sampled k-space area of whole contrast sets. The contrast-specific edge map (W) in the step 2) was estimated by solving the following least squares problem:
$$\arg \underset{u_{i}}{min}\left \| v_{i}-Eu_{i} \right\|_{2}^{2}+\lambda \left \| J(W)(u_{i}-W))) \right \|_{2}^{2}$$
Finally, multi-contrast images were reconstructed by solving the following optimization problem:
$$\arg \underset{x_{i}}{min}\left \| y_{i}-Ex_{i} \right\|_{2}^{2}+\lambda \left \|u_{i}^{*}\partial x_{i} \right \|_{1}^{}$$
Where i is the index of each contrast (i = 1,2,3 for T1, T2, FLAIR, respectively), vi is measured k-space gradient, yi is the measured data in k-space, E = C i Fu is the encoding operator, λ is the regularization parameter, ||∙||2 is the Euclidian norm, and ||∙||1 is the 1-norm. Figure 1 represents a schematic of the proposed method.
Results
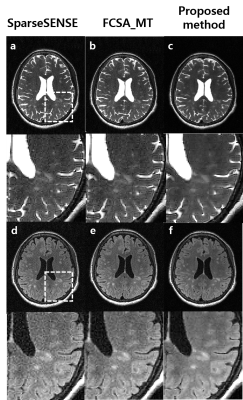
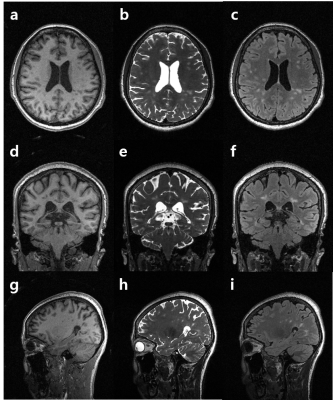
Figure 2 represents T2- and FLAIR T2-weighted brain images with MS lesions reconstructed using conventional SPARSE SENSE (a) [3], FCSA_MT (b) [4], and the proposed method (c) for comparison (R = 6). An ROI , which includes MS lesions, is zoomed in. The proposed method clearly depicts MS lesion without an apparent artifacts and amplified noise even at a high R = 6. Figure 3 shows multi-contrast 3D images reconstructed using the proposed method and then reformatted into the axial, coronal, and sagittal orientations. The proposed method yields high resolution 3D multi-contrast images roughly in 5 minutes, successfully delineating MS lesions.Discussion and Conclusion
We successfully demonstrated that highly accelerated, multi-contrast 3D images can be produced over the whole brain roughly in 5 minutes without an apparent artifact and amplified noise from patients with MS lesions. This proposed method strengthened the contrast between lesions and normal tissue and clearly depicted lesions characteristics on basis of multiple parameters. It can accurately detect small lesions and quantify lesions features for further analysis. Therefore, multi-contrast 3D images and derived quantitative characteristics have potential capability for early precise diagnosis and minitor patients’ conditions and then as imaging biomarkers for optimal management in clinical practice.Acknowledgements
No acknowledgement found.References
[1] Polman, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the ‘‘McDonald Criteria.’. Ann Neurol 2005;58:840–846
[2] Greg Ongie, et al. Off-the-grid recovery of piecewise constant images from few fourier samples. CoRR. 2015; abs/1510.00384
[3] Otazo, et al. Combination of Compressed Sensing and Parallel Imaging for Highly Accelerated First-Pass Cardiac Perfusion MRI. MRM. 2010; 64:767–776
[4] Junzhou Huang, et al. Fast multi-contrast MRI reconstruction. MRI. 2014; 32:1344–1352
Figures