1176
Non-rigid Brain MRI Registration Using Two-stage Deep Perceptive Networks1School of Automation, Northwestern Polytechnical University, Xi'an, China, 2Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 3School of Biomedical Engineering, Med-X Research Institute, Shanghai Jiao Tong University, Shanghai, China
Synopsis
Non-rigid image registration is a fundamental procedure for the quantitative analysis of brain images. The goal of non-rigid registration is to obtain the smooth deformation field that can build anatomical correspondences among two or more images. Conventional non-rigid registration methods require iterative optimization with careful parameter tuning, which is less flexible when dealing with the diverse data. Therefore, we propose a two-stage deep network to directly estimate the deformation field between an arbitrary pair of images. This method can tackle various registration tasks, and is consistently accurate and robust without parameter tuning. Thus, it is applicable to clinical applications.
Purpose
We propose to learn a general deep network [1], to directly map from the input image pair (e.g., an arbitrary pair of fixed and moving images) to their final deformation field. Then, in the application stage, the deformation field of an unseen image pair can be effectively and accurately obtained through the learned deep network without tedious parameter tuning. Since the mapping between the image pair and their deformation field is complex and highly non-linear, we propose a two-stage deep perceptive network, which can solve the non-rigid registration problem from scratch.Method
Basically,
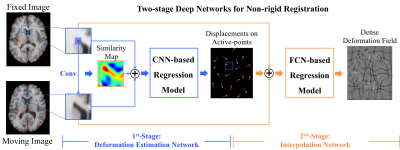
the proposed two-stage deep perceptive network in Fig. 1 is designed in a
patch-wise manner. The 1st-stage is a deformation estimation network, which estimates
the local deformations of the selected active patches/points. The 2nd-stage
is an interpolation network, which
can generate the dense deformation field based on those irregularly distributed
active points and their deformations.
The
1st-stage
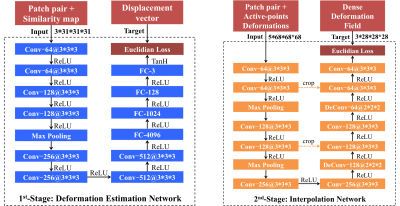
is designed based on a convolutional neural network (CNN), where the input is
the patch pair extracted from the same center in both the fixed and the moving
images. The desired output is the displacement vector of the center location.
In this stage, the center locations of the training samples (i.e., the patch
pairs) are the active points, which are associated with high image gradients [2]
and have abundant appearance saliency. Thus, the sampled patches can be more
informative, which can help more accurate correspondence matching and local
deformation estimation. We further propose to incorporate the contextual cue,
i.e., the local similarity between the pair of input patches, to enhance
perceptive capability of the network. The contextual cue and the pair of input
patches are concatenated together to feed to the CNN-based regression model for
learning the corresponding displacement vector.
The 2nd-stage is a fully convolutional network (FCN) based interpolator. It can effectively interpolate the dense deformation field via those irregularly distributed active points, rather than using the common spline-based interpolator that is time-consuming and less accurate. Specifically, the input is the displacement estimations of the sparse active points as well as the pair of image patches that provide contextual information. The output is the dense deformation field as a whole.
The
two networks are trained independently for different goals in the training
phase (c.f., Fig. 2), and then jointly used in the application phase to
complete the registration. In the application phase, the learned deep network
in the 1st-stage is used
to predict the displacement vectors at the active points that sufficiently
cover the whole brain volume. The final deformation field is immediately obtained
via the 2nd-stage
interpolation network.
Results
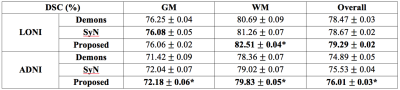
We have validated our proposed method on two different datasets, i.e., LONI (40 subjects) and ADNI (20 subjects), which cover both the young and old adult brain MR images. We train the deep network using 30 subjects in LONI and test on the remaining 10 subjects. We also directly apply the model trained from LONI to the challenging ADNI dataset, which often have large local deformations across subjects. Demons [3] and SyN [4] are used as the comparison methods.
The registration performances in term of Dice Similarity Coefficients (DSC) [5] are reported in Table 1. For both datasets, the proposed method can achieve the best overall performance, especially for the challenging ADNI dataset. Note that, our method uses only 1% of the whole image voxels as the active points to generate the whole deformation field, without iterative optimization and parameter tuning. This well demonstrates the generalization of the trained model. Also, our proposed registration method is robust and accurate for diverse registration tasks, which makes it more flexible and applicable in practice.
Discussion and Conclusion
We propose a novel deformable registration method that uses the two-stage deep network to directly establish the complex mapping from an image pair to their corresponding deformation field. To effectively solve the registration problem, we first employ a conventional CNN model to estimate the deformations of the active points that possess rich anatomical details, and then efficiently generate the dense deformation field through the FCN-based interpolation network. Experiments on two different datasets show the consistent high performance of our proposed method, indicating its potential applications to various clinical scenarios.Acknowledgements
No acknowledgement found.References
- Shen, D., G. Wu, and H.-I. Suk, Deep Learning in Medical Image Analysis. Annual Review of Biomedical Engineering, 2017(0).
- Cao X, Yang J, Zhang J, et al. Deformable Image Registration Based on Similarity-Steered CNN Regression[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2017: 300-308.
- Vercauteren, T., et al., Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage, 2009. 45(1): p. S61-S72.
- Avants, B.B., et al., Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis, 2008. 12(1): p. 26-41.
- Cao X, Yang J, Gao Y, et al. Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis[J]. Medical Image Analysis, 2017.
Figures