1168
Simultaneous detection of cardiac, respiratory, and rigid body head motion using the scattering of a parallel transmit RF coil at 7T1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, OXFORD, United Kingdom, 2University of Oxford Centre for Magnetic Resonance Research, University of Oxford, OXFORD, United Kingdom, 3Oxford Centre for Human Brain Activity, Wellcome Centre for Integrative Neuroimaging, Department of Psychiatry, University of Oxford, Oxford, United Kingdom, 4British Heart Foundation Centre of Research Excellence, Department of Cardiovascular Medicine, Oxford, OXFORD, United Kingdom
Synopsis
Cardiac pulsation, respiration and rigid-body head motion induce changes in relative tissue conductivity. These changes can be detected via the scattering of a parallel transmit RF coil array at 7T, by monitoring the forward and reflected power of each channel, with a 5ms pulse. The time series of the scattering data can be used to detect cardiac and respiratory motion, as well as predict head position using training data. We show that detected heart and respiration rates match those recorded with independent monitoring, and head position may be predicted with reasonable precision, at little extra time cost.
Purpose
To demonstrate the viability of monitoring rigid-body head motion, cardiac pulsation and respiration by measuring the scattering of a multi-channel pTx head coil.Introduction
Cardiac pulsation, respiration and rigid-body head motion all induce changes in relative tissue conductivity. We hypothesise that these changes can be detected by measuring the forward and returned voltages of a parallel transmit head coil1, allowing for the monitoring of such motions at little time cost.Methods
All imaging was performed on a Magnetom 7T scanner (Siemens, Erlangen, Germany) equipped with an 8Tx/32Rx Nova Medical Head coil with local SAR monitoring. Volunteers A (35,F) and B (30,F) were instructed to move to different head positions by rotating their heads around the left-right or head-foot axes, by moving their heads in 3 steps for right/left/pitch-up and 2 steps pitch-down directions, before returning to the original centre position. For each position, a 3D GRE image (FOV=280x280x176mm3, matrix size: 128x128x88, TR/TE=3.5/1.51ms, FA=15deg ) and a scattering matrix (S-matrix) were acquired. S-matrices were measured using a frequency multiplexed Gaussian RF pulse (channel spacing: 2kHz) of 5ms, played out with a TR of 10/20ms for volunteers A and B, and calculated from the forward ($$$V_{fwd}$$$) and returned ($$$V_{ret}$$$) voltage detected by the directional couplers of each of the eight transmit channels: $$$V_{ret}=S_{i,j} \cdot V_{fwd}$$$. In the first, central position, a baseline time series of S-matrices was acquired over 6000TRs. For each following position, a short S-matrix acquisition of 200TRs was played out. Volunteer A was instructed to perform small head motions (1-2mm range), volunteer B was instructed to perform large head motions (5-10mm range). For volunteer A, heart rate and respiration were recorded with a modular data acquisition system (MP150, BIOPAC Systems Inc., USA) using a pulse oximeter and a breathing belt, without synchronisation to the MRI acquisition. All data was analysed using MATLAB (Mathworks, Natick, USA) and bespoke tools. Cardiac and respiratory motion information was extracted from the 6000 time-point series of the 8-by-8 complex S-matrices acquired in the first position on volunteer A. After temporal de-trending, ICA algorithm was used to separate signal components2. Power spectrum analysis was used to determine the cardiac and respiratory components, defined as the highest power signals in frequency bands of fcardiac=0.6-2.4Hz and fresp=0.08-0.33Hz. Peak detection was used to detect the individual R-R intervals of cardiac motion, and the breathing rate. The respiratory component was smoothed with a sliding window of 100ms to aid peak detection. The detected heart and breathing rates were compared to the gold standard of MP150 data. Relative head position was estimated from image data via image realignment, using SPM12. Head position was estimated from the S-matrix data using general linear models. The 128 components (real and imaginary) of the S-matrix were reshaped as a vector ($$$n_{Smat}$$$), and used to predict six degrees of motion derived from realignment: $$$n_{motion}=n_{Smat} \cdot \beta $$$. To asses the robustness of S-matrix data, two models were used, utilising the first ("coarse") or the time-average of all ("fine") S-matrices of a position. Tikhonov regularisation was used to avoid overfitting, with lambda empirically set to 100 times the standard deviation of a predictor over the 6000 acquisitions of the S-matrix in the first position. Performance of the model was tested by predicting one head position using a model constructed from all other head positions.Results
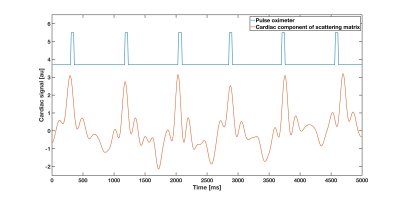
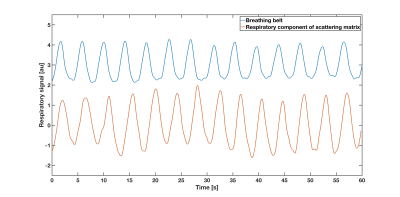
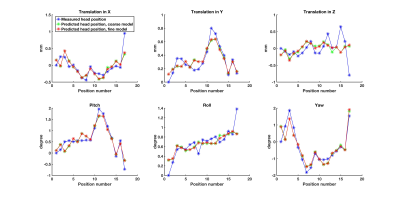
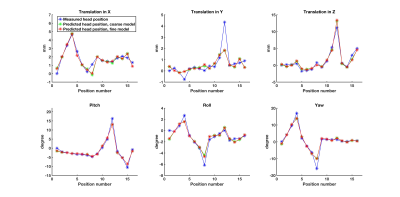
Peak intervals for cardiac and respiratory motion estimated from S-matrix data (861±37ms; 4235±288ms) were closely matched to the gold standard (877±40ms; 4212±264ms). The two cardiac/respiratory signals are shown in Figs.1/2. Small head motions of volunteer A were predicted with a mean prediction error across all positions of 0.1/0.1/0.2mm for x,y,z and 0.2/0.1/0.3deg for pitch, roll, yaw for both models (Fig.3). For the large motions of volunteer B, the mean prediction errors were 0.3/0.4/0.6mm and 0.8/0.5/1deg for x,y,z and pitch, roll, yaw, for both models (Fig.4), indicating good performance and robustness for even the coarse model.Discussion
Cardiac pulsation and respiration were successfully detected, in line with previous results in cardiac imaging1,2. Head position was estimated with reasonable precision for both large and small head motions, using a trained model. The proposed method requires little additional scan time, with head position estimated from just 5 ms of data, and can be easily deployed on any pTx coil. Further work will look at using scattering coefficients, which require no sequence modification1 for data collection, and methods that require no training for motion prediction.Conclusion
We have demonstrated the feasibility of detecting both pulsatile and rigid-body head motion from scattering matrix data collected by a pTx coil.Acknowledgements
The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z).References
1: Hess AT et al: Diaphragm position can be accurately estimated from the scattering of a parallel transmit RF coil at 7T, Magn Reson Med, 2017
2: Jeaschke S et al: Contact-free cardiac motion estimation using the scatter of a parallel transmit coil t 7T, Proc. Intl. Soc. Mag. Reson. Med. 25 3262, 2017
Figures