1136
Pulseq-GPI: A single, comprehensive framework for MR method developmentSneha Potdar1, Pavan Poojar1,2, Ashok kumar Reddy2, Keerthi Sravan1,3, Stefan Kroboth4, Jon-Fredrik Nielsen5, Maxim Zaitsev4, Ramesh Venkatesan2, and Sairam Geethanath1,6
1Medical Imaging Research Center, MIRC, Dayananda Sagar Institutions, Bengaluru, India, 2Wipro GE Healthcare Pvt Ltd, Bengaluru, India, 3New York University Tandon School of Engineering, New York, NY, United States, 4Dept of Radiology, Medical Physics, Medical Center University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany, Freiburg, Germany, 5Functional MRI Laboratory, University of Michigan, Michigan, MI, United States, 6Dept. of Radiology, Columbia University Medical Center, New York, NY., Columbia, SC, United States
Synopsis
This work develops and integrates an open source implementation of the
Purpose
To demonstrate a unified MR method development framework comprising of sequence implementation (and simulation), deployment, image reconstruction, and analysis.Introduction
Pulseq1 enables design of pulse sequences in MATLAB (The Mathworks Inc., MA) programming environment that can currently run on three vendor hardware platforms. Graphical Programming Interface2 (GPI) is an open source tool for developing pulse sequences based on Python. This work aims to simplify the process of designing MR pulse sequences by integrating Pulseq’s functionality with the GPI. In the proposed platform, “Pulseq-GPI”, pulse sequences are developed via a simple drag and drop interface.Methods
The mr toolbox in its entirety was rewritten in Python 3.6.1 as the ‘mr_GPI’ package3 and integrated with GPI as nodes. Three implementations of Pulseq (MATLAB, Python and Pulseq-GPI) were employed and demonstrated via an implementation of the Spin Echo (SE) sequence. In vitro phantom scans of the American College of Radiology MRI phantom4 were acquired on a GE 1.5T scanner utilizing the TOPPE interpreter5. The scans were performed using an eight-channel head coil. The SE pulse sequence was implemented in Python and GPI with identical sequence parameters as in the MATLAB implementation: maximum gradient amplitude 33mT/m, maximum slew rate 100T/m/s, field of view 256mm, matrix size 256x256, TR/TE=2000/100ms. The sequence was also implemented in Python-Pulseq and Pulseq-GPI, and plots were obtained. In this particular implementation, the acquisition parameters were fed to the Pulseq-GPI sequence network (spin_echo.net) available online6. The .seq file generated was fed to the interpreter block. The scripts in this block converted the .seq file contents to vendor specific files (seq2ven; TOPPE in this case for the GE scanner) along with plotting the waveforms to be verified on the vendor software. Upon verification of the waveforms, the sequences were run on the scanner. The raw data generated by the scanner was then reconstructed using GPI’s inbuilt FFT_NUMPY. All reconstructions involved a magnitude sum-of-squares channel combination. The implementation code execution times of the mr toolbox and Pulseq-GPI for SE were measured and compared over three trials. The MATLAB and Python implementations of the sequences were also evaluated based on the number of lines of code. The abilities of Pulseq-GPI as a comprehensive tool for algorithm development were also tabulated in comparison with other open source tools.Results
Figure 1 shows the workflow of the SE sequence as an example using Pulseq-GPI -- from sequence design to image analysis. More importantly, the figure demonstrates the open-source, comprehensive pipeline for MR method development that Pulseq-GPI was intended for. It can be observed that three (light gray) out of the five (or more) blocks are vendor independent. The network for the implemented SE sequence is shown in Figure 2. It can be noticed that the sequence can be assembled and written to ‘.seq’ format using only four types of nodes. The resulting waveforms from MATLAB-Pulseq and Pulseq-GPI are shown in figure 3 (Python-Pulseq provides identical waveforms as Pulseq-GPI invokes Python-Pulseq). It can be observed that the waveforms are identical. Figure 4 depicts the images acquired from the vendor sequence and Pulseq-GPI along with line intensity profiles. It can be observed that the two images have a minute pixel shift. This may be attributed to changes in the reconstruction pipelines of the two implementations. It can also be observed the signal-to-noise ratio of the vendor implementation is comparatively better then Pulseq-GPI. This may be either attributed to an imperfect implementation of the refocusing pulse in Pulseq-GPI or the use of raw data filtering in the vendor implementation which has not been performed in the case of Pulseq-GPI. The computation time for SE in Pulseq-GPI is approximately 18± 0.1s when compared with MATLAB-Pulseq which takes around 3.170 ± 0.01s, and Python-Pulseq which takes around 6.582 ± 0.2s. The slower computation times of Pulseq-GPI was primarily attributed to the multiple back and forth calls between the node UI and the code.Discussion and Conclusion
Pulseq-GPI provides two major benefits that have been demonstrated. Firstly, a unified framework for MR method development research related to simulations, pulse sequence design and implementation, image reconstruction and image analysis. Secondly, it offers the MR researcher two options to develop and/or implement MR algorithms based on his/her programming ability: Python-Pulseq for the enthusiastic programmer and Pulseq-GPI for the researchers focused on rapid prototyping. Table 1 illustrates a few of the comparative advantages of Pulseq-GPI over other open source frameworks. Pulseq-GPI is completely open source given that it has been programmed in Python. Current and future work includes incorporation of Specific Absorption Ratio and Peripheral Nerve Stimulation evaluations to include safety validations.Acknowledgements
Ministry of Electronics and Information Technology, “National Mission on Indigenous MRI” 1(15)/2014-ME&HI, Department of Science and Technology (DST), grant# DST/TSG/NTS/2013/100-G and DST/VGST/KFIST/LII/GRD333 for support.References
1. Layton, K.J. et. al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magnetic resonance in medicine, 2017; 77(4), pp.1544-1552. 2. Zwart, Nicholas R., et al. Graphical programming interface: a development environment for MRI methods. Magnetic resonance in medicine 2015;74, no. 5: 1449-1460. 3. Keerthi Sravan R, et. al. Implementation of Pulseq in GPI Lab. 2017; ISMRM 4 Chen, C.C., et. al. Quality assurance of clinical MRI Scanners using ACR MRI phantom: preliminary results. Journal of digital imaging, 2004;17(4), pp.279-284. 5. https://toppemri.github.io/ 6. https://github.com/mirc-dsi/IMRI-MIRC/tree/master/PSDFigures
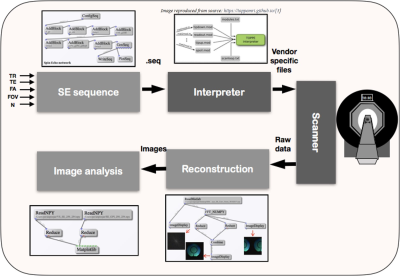
Figure 1:Pipeline for SE sequence as an example of comprehensive algorithm developed. Dark gray represent the vendor specific components while blocks in light gray are vendor independent.

Figure 2: Spin Echo network for Pulseq-GPI with the UI
for sequence parameters.
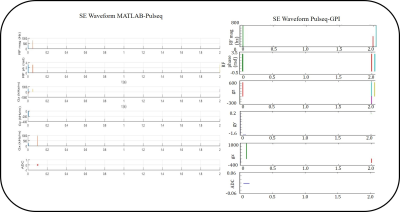
Figure 3: Pulse Sequence timing diagram for SE obtained from MATLAB-Pulseq and Pulseq-GPI for 1 TR – 2s.
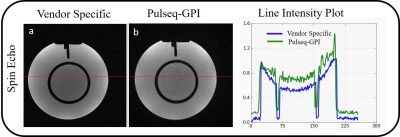
Figure 4: Reconstructed images using GPI network (a) is Vendor specific image and (b) is Pulseq-GPI image. The vertical red line shows the pixels used for plotting line intensity plot (c).
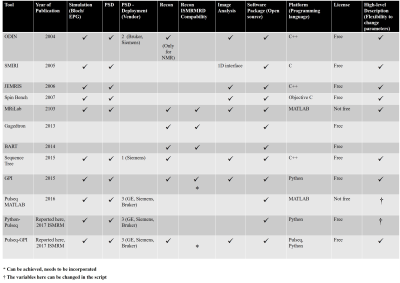
Table 1:
MR software platforms compared with Pulseq-GPI, with respect to different parts of the MR pipeline.