1097
GPU-based Monte-Carlo simulation of diffusion in astrocytes reconstructed from confocal microscopy1Molecular Imaging Research Center (MIRCen), Commissariat à l'Energie Atomique, Fontenay-aux-Roses, France, Fontenay aux Roses, France
Synopsis
Here we implement a GPU-based Monte-Carlo simulation of diffusion to efficiently simulate diffusion-weighting in realistic, complex cellular structures such as astrocytes as directly derived from confocal microscopy. This opens new possibilities to better understand intracellular diffusion, validate diffusion models, and create dictionaries of intracellular diffusion signatures.
Purpose
The aim of this works is to develop efficient Monte-Carlo simulation of diffusion in complex cellular structures, such as astrocytes as directly derived from confocal microscopy, to calculate diffusion-weighted NMR signal in the intracellular space.Methods
Triangular surface mesh of astrocyte: Mouse brain slices were acquired using a Leica SP8 confocal system and confocal image resolution was 0.07443×0.07443×0.2985 µm3. The volume image of mouse's astrocytes was extracted from the confocal microscopy images, and was then used for generating the triangular surface mesh by using an open-source meshing toolbox "Iso2Mesh"1 adapted with the cgalmesh2 library. Typical astrocytic surfaces consist of 5×105 - 1×106 faces.
GPU-based Monte-Carlo simulation: Random walk of particles (N = 217) was simulated inside the structure with free intracellular diffusivity Dintra = 0.5 µm2/ms and 5000 time-step. We implemented a rejection-sampling method whereby the particle position is updated only if the new position is inside the astrocyte. To manage (and accelerate) the interaction of the particles with the astrocyte's surface, we used an octree structure3 to identify faces that particles can encounter during the next time step. Octree is a hierarchical data structure based on a recursive spatial space decomposition of a 3D data. The root node is represented by a cube containing whole data. Each node containing more than 32 data points (or some other value, depending on data's size) is divided into eight children (octant). The astrocyte's surface triangular mesh is described by the circumcenter points and the circumradii. The circumcenter points are used as data to build the octree structure and the circumradii information are used in the octree search algorithm. Diffusion-weighted signal was computed using the phase accumulation approach (as described in4,5).
All codes were implemented in C++ using the CUDA library to interface with NVIDIA GPUs (Tesla K40c) and performed on an HP workstation (Intel(R) Xeon(R) CPU E5-2630 v4 @ 2× 2.20GHz) on Windows 7 professional.
Results
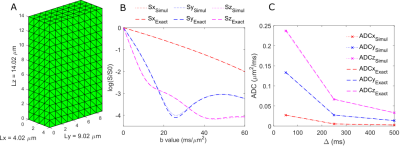
First of all, we validated the numerical simulation of the diffusion signal attenuation, as well as the ADC (calculated as $$$\frac{-ln(S(b=3))}{3}$$$) as a function of ∆, in the simple case of restricted diffusion in the box of size Lx=4.02, Ly=9.02 and Lz=14.02 µm (fig. 1A). The good agreement between the simulated signal attenuation and analytical signal attenuation6 in the case of infinite short gradient duration (δ=0 ms), diffusion time ∆=50 ms and very high b-values (b from 0 to 60 ms/µm2) is shown on fig. 1B. Fig. 1C showing the superimposed between the simulated and analytic ADC behavior when increasing the diffusion time from 50 ms to 500 ms.
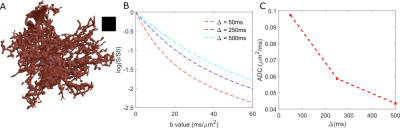
Moreover, we ran the simulation for diffusion in astrocyte reconstructed from confocal microscopy image (fig. 2A) for three different diffusion times (∆=50, 250 and 500 ms) and high b-values (up to 60 ms/µm2). The simulated signal attenuation is shown in fig. 2B. The corresponding ADC (calculated as $$$\frac{-ln(S(b=3))}{3}$$$ ) as a function of ∆ is shown in fig. 2C.
Simulated data can then be analyzed using existing models, which might be interesting to investigate the validity of these models. For example, it is possible to fit the high-b data simulated at ∆=50 ms using an analytical model of diffusion in randomly oriented cylinders of infinite length but finite diameter, as we introduced for experimental intracellular metabolite diffusion measured by diffusion-weighted MRS7. If we do so, we extract Dintra=0.31 µm2/ms and cylinder diameter d~3.4 µm. The lower Dintra compared to ground truth (0.5 µm2/ms) is certainly due to the presence of many secondary structures along the main astrocytic processes8, while the "ground truth" fiber diameter as measured for this astrocyte using VAA3D reconstruction software9 is ~4.7 µm, which is in reasonable agreement with the simple cylinder model, but suggests that there is still room to improve models to analyze intracellular diffusion at high-b. Very interestingly, these values for Dintra and d are very close to values extracted from experimentally measured myo-inositol diffusion7 (Dintra = 0.32 µm2/ms and d~3.1 µm), which is supposed to be an astrocytic metabolite.
Discussion and conclusion
The successful implementation of GPU-based Monte-Carlo diffusion simulation opens the possibility to simulate diffusion-weighting in complex cellular structures, which may help better understand intracellular diffusion, validate diffusion models, and create dictionaries of intracellular diffusion signatures.Acknowledgements
This work was funded by the European Research Council grant number 336331 (INCELL project). The Tesla GPU was generously donated by NVIDIA Corporation.References
- Qianqian Fang and David Boas, Tetrahedral mesh generation from volumetric binary and gray-scale images, Proceedings of IEEE International Symposium on Biomedical Imaging, 2009, pp. 1142-1145
- CGAL, Computational Geometry Algorithms Library, http://www.cgal.org
- Donald Meagher, Geometric modeling using octree encoding, In Computer Graphics and Image Processing, Volume 19, Issue 2, 1982, Pages 129-147, ISSN 0146-664X, https://doi.org/10.1016/0146-664X(82)90104-6.
- Hall, M. G. and Alexander, D. C., Convergence and Parameter Choice for Monte-Carlo Simulations of Diffusion MRI, IEEE Transactions on Medical Imaging, 2009, 28, 1354-1364
- Waudby, C. A. and Christodoulou, J., GPU accelerated Monte Carlo simulation of pulsed-field gradient NMR experiments, Journal of Magnetic Resonance, 2011, 211, 67 - 73
- Tanner, J. E. and Stejskal, E. O. Restricted self-diffusion of protons in colloidal systems by the pulsed-gradient, spin-echo method, The Journal of Chemical Physics, AIP, 1968, 49, 1768-1777
- Palombo, M.; Ligneul, C. and Valette, J., Modeling diffusion of intracellular metabolites in the mouse brain up to very high diffusion-weighting: Diffusion in long fibers (almost) accounts for non-monoexponential attenuation, Magnetic Resonance in Medicine, 2017, 77, 343-350
- Palombo, M.; Ligneul, C.; Hernandez-Garzon, E. and Valette, J., Can we detect the effect of spines and leaflets on the diffusion of brain intracellular metabolites? NeuroImage, 2017
- VAA3D, 3D Visualization-Assisted Analysis, www.vaa3d.org
Figures