1062
Dynamically Switched B0 Field Control for Separate Optimization of Tailored Volume Lipid Suppression and B0 Homogeneity for Brain Chemical Shift Imaging at 3T using Multi-Coil Shim Array1Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospita, Charlestown, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States, 4Fetal Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 5Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, United States
Synopsis
A rapidly reconfigurable 32-channel local-multi-coil-shim-array is used
to both enhance lipid suppression and narrow metabolite linewidth in
chemical-shift imaging of the brain. Using in-situ optimization, the
array is first configured to widen the spectral gap between spatially
separate lipid and metabolite regions, to improve lipid-surpressing
inversion, and then reconfigured for field homogeneity, to narrow
metabolite linewidth during readout. For 2cm thick brain slab, using
the dynamically-reconfigured array reduced lipid contamination by 24.5%,
reduced linewidth by 34%, and increased well-imaged brain area by 38% over static 2nd order shimming
Introduction
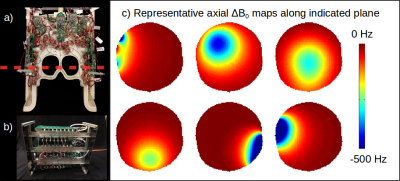
Lipid suppression poses a serious challenge for magnetic resonance spectroscopic imaging (MRSI) of the brain. Common lipid suppression methods exploit either the spectral or spatial separation of metabolites and lipids, including frequency selective inversion1,2 or spatially selective saturation methods2-7. New in this work is switching between tailored-volume lipid suppression and homogeneity enhancement, by using a rapidly-reconfigurable a multi-coil (MC) array8,11. The array, 32 small coils in close proximity to the subject, shown in figure 1, is first used to produce spatially varying static fields that separate lipids from metabolites. Greater lipid-metabolite separation improves frequency selective inversion performance, exploiting both the spectral and spatial separation of lipids from metabolites. Enabled by the robust switching of MC arrays, field control is switched from lipid metabolite separation to brain homogenization for metabolite imaging. Rapid reconfiguration of static fields improves lipid suppression without compromising the optimal shim for metabolite readout.
Complete lipid inversion without impacting metabolites requires the chemical shift in the brain volume to be outside of the inversion and transition band of the spectrally selective RF pulse simultaneously ensuring the lipid shifts are restricted within the inversion band. MC Array currents are calculated by minimizing the sum of the violation of the transition band constraint:
$$\min_\text{currents}\sum\text{overlap}\\\text{ s.t. }\text{NAA}-\text{overlap}\ge\max(\text{lipid})-\text{transition};\text{overlap}\ge0$$
Methods
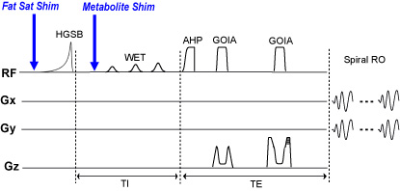
MRSI brain-slab datasets are acquired with a baseline 2nd order shim, volume-tailored lipid suppression, and tailored lipid suppression with homogeneity shimming using an adiabatic spin-echo sequence using an AHP-HS8 excitation pulse, WET water suppression pulses, and spiral readout2. Fat suppression uses a hypergeometric single band asymmetric adiabatic inversion pulse with a sharp transition band of 70 Hz and an inversion band of 2 kHz2. The transition band is positioned at 1.7ppm between the NAA peak at 2ppm and the dominant lipid peak at 1.3ppm. Transition band position is adjusted accordingly during the lipid saturation experiments. Figure 2 shows the pulse sequence and the time of triggers used to update the MC array current settings. Acquisition parameters included TR/TE=1600/97 ms, FOV=240x240 mm, matrix size 24x24, slice thickness of 2 cm, 6 averages, TA = 1:30 min. MRSI data is zerofilled in k-space to 32x32, Hamming filtered and fitted with LCModel.
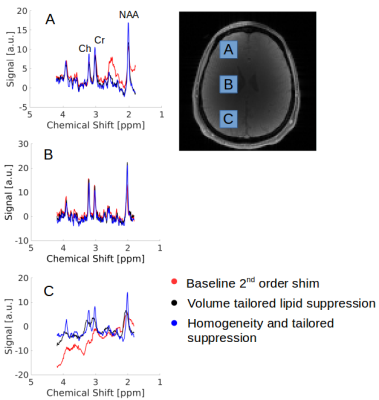
Lipid, spectral linewidth, metabolic maps, and spectra from three different brain regions (frontal, mid-brain, occipital) are compared between the methods. The mean lipid contamination, mean linewidth and the number of acceptable voxels are compared quantitatively. Lipid contaminations are calculated by summing spectral magnitudes in the range from 1.9ppm to 0.8ppm. The spectral linewidth is computed by LCModel, and a voxel is defisned acceptable if its SNR>5, linewidth<15 Hz, Cramer Rao-Lower bounds of tNAA<25%, and a ratio of NAA to Creatine<2. The latter excludes voxels with strong lipid contamination fitted by LCModel as NAA.
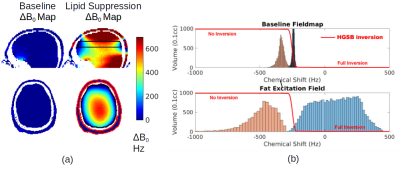
Figure 3a shows a baseline field map and calculated MC-generated fields for tailored-volume lipid suppression. Figure 3b, a histogram of chemical shifts within each region, shows the widened gap between metabolite and fat spectra that insures comprehensive inversion of the fat, but also produces an undesirable broadening of the metabolite spectrum. Reconfiguring the MC array after lipid suppression, to homogenize ΔB0 results in narrow metabolite spectra during water suppression and readout. In either configuration, coil currents are less than 3.5A and the array total is less than 50A.
Results
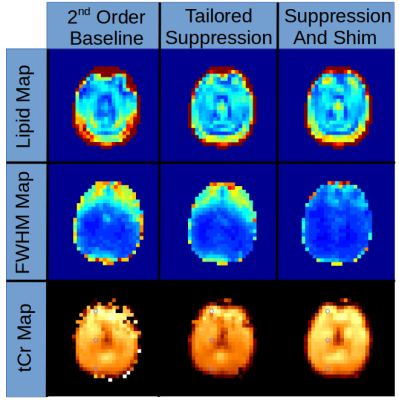
Figure
4 shows the improvements in lipid suppression and spectral quality
achieved using the dynamically-switched MC array compared to the
static-2nd-order baseline B0 shim. Mean lipid
signal over the entire 2cm slab was reduced by 27.7 %, while
tailored-volume lipid suppression and homogeneity shiming reduced the
fat by 24.5 % in comparison to the static scanner-shim.
Lipid maps show
significantly reduced lipid contamination in posterior and anterior
regions of brain near the skull Homogeneity shimming improved the
mean FWHM line-width from 10.9Hz to 7.2Hz (34%). FWHM maps show voxel
T2* improvement from homogeneity shimming especially in the region
above the sinuses. Scanner
baseline 2nd
order shim MRS data
63.9%
brain voxels were
acceptable, with
the addition of tailored
lipid suppression 72.7%.
With homogeneity
shimming and tailored
lipid suppression 88.3% of
voxels were acceptable.
Discussion
A time-divided pulse sequence with distinct optimal static fields satisfying different objectives, when combined with fast switching of high spatial order array currents, enables time-divided static field control functions for improved imaging performance. The high spatial order of MC array fields provides a useful static field basis for both tailored-volume lipid suppression and B0 homogeneity shimming within the brain. Sequence acquisition time and SAR are not adversely impacted, as the method required no modification of the pulse sequence. Subject specific calibration is fast requiring only the acquisition of a baseline B0 map.Acknowledgements
Funding support from NH K99EB021349 and MITEECS Shillman FellowshipReferences
- Balchandani P, Spielman D. Fat suppression for 1H MRSI at 7T using spectrally selective adiabatic inversion recovery. Magnetic resonance in medicine. 2008 May 1;59(5):980-8.
- Esmaeili M, Bathen TF, Rosen BR, Andronesi OC. Three‐dimensional MR spectroscopic imaging using adiabatic spin echo and hypergeometric dual‐band suppression for metabolic mapping over the entire brain. Magnetic resonance in medicine. 2017 Feb 1;77(2):490-7.
- Star‐Lack J, Nelson SJ, Kurhanewicz J, Huang L, Vigneron DB. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING). Magnetic resonance in medicine. 1997 Aug 1;38(2):311-21.
- Boer VO, van de Lindt T, Luijten PR, Klomp DW. Lipid suppression for brain MRI and MRSI by means of a dedicated crusher coil. Magnetic resonance in medicine. 2015 Jun 1;73(6):2062-8.
- Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H‐MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magnetic resonance in medicine. 2008 Jan 1;59(1):1-6.
- Luo Y, De Graaf RA, DelaBarre L, Tannus A, Garwood M. BISTRO: an outer‐volume suppression method that tolerates RF field inhomogeneity. Magnetic resonance in medicine. 2001 Jun 1;45(6):1095-102.
- Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer‐volume suppression saturation bands. Journal of Magnetic Resonance Imaging. 2011 Apr 1;33(4):792-802.
- Stockmann JP, Witzel T, Keil B, Polimeni JR, Mareyam A, LaPierre C, Setsompop K, Wald LL. A 32-channel combined RF and B0 shim array for 3T brain imaging. Magn. Reson. Med. 2016;75(1):441-51.
- Juchem C, Rudrapatna SU, Nixon TW, de Graaf RA. Dynamic multi-coil technique (DYNAMITE) shimming for echo-planar imaging of the human brain at 7 Tesla. Neuroimage. 2015 Jan 15;105:462-72.
- Kizildag E, Stockmann P, Gagoski B, Guerin B, Grant E, Wald L, Elfar A. Improved spiral chemical shift imaging at 3 Tesla using a 32-channel integrated RF-shim coil array. Int. Soc. Magn. Res. Med. 2016, p.0380
- Arango N, Stockmann JP, Witzel T, Wald L, White J. Open-source, low-cost, flexible, current feedback-controlled driver circuit for local B 0 shim coils and other applications. Int. Soc. Magn. Res. Med. 2016, p. 1157.
Figures