1051
Automatic Segmentation of Carotid Vessel Wall in Multi-Contrast Blackblood Images using Deep Learning1Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China, 2Vascular Imaging Laboratory, Department of Radiology, University of Washington, Seattle, WA, United States
Synopsis
In this work, we proposed an automatic approach for segmentation of carotid vessel wall in multi-contrast blackblood images, using a fine-tuning U-net neural network. The U-net network consists of an encoder path that captures context and reduces data dimension and a symmetric decoder path that enables precise localization and high resolution. The fine-tuning was utilized to accommodate multi-contrast images input. The pixel-level sensitivity, specificity and IoU of our model achieved 0.869, 0.987 and 0.751 on the test data set, respectively.
Introduction
It is of crucial importance to estimate whether there is an atherosclerosis in carotid artery through the information of carotid vessel wall images. However, it is time-consuming and subjective for radiologists to segment the carotid vessel wall in multi-contrast MR images. In this work, we aimed to develop an automatic approach for segmentation of carotid vessel wall in multi-contrast blackblood images based on U-net network.Methods
Our data set includes 1,098 subjects (16 locations per subject, and totally 17,568 locations) with institutional review board approval. Each location has four contrasts1 (T1, T2, TOF and MPRAGE) with 256*256 in image size. All data were viewed by experienced radiologist using CASCADE2. Out of the 1,098 subjects, we used 660 for training, 219 for validating and 219 for testing.
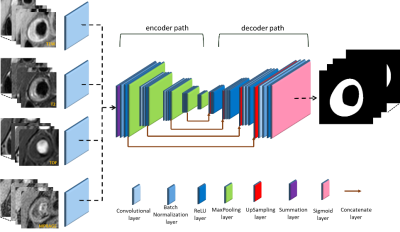
The segmentation of carotid vessel wall in multi-contrast MR images was formulated as a pix-wise binary classification problem. However, we need to obtain enough information of vessel wall as well as keep high image resolution. In the U-net network3 (Figure 1), the encoder path consists of standard 2D convolutional layer, batch normalization (BN) layer, and rectified-linear non-linear (ReLU) layer followed by a max-pooling layer. This encoder network reduces the data dimension from 256*256 to 8*8 and obtains high-level features. The decoder path consists of up-sampling layers and concatenate layers to recover high resolution pixel-wise labels4. After the operation of the decoder network, the data dimension increases from 8*8 to 256*256. The final layer of decoder network is a binary sigmoid classifier that produces probability of carotid vessel wall for each individual pixel. In particular, we made four copy of the first convolution layer in the original U-net network and connected each MRI sequence image with one copy. Then we merged the results of these four convolution layers by summation, and connected to the rest of the net. The full segmentation algorithm is summarized in Figure 1. In addition, we flipped the image horizontally at 0.5 probability level and cropped the image randomly as data augmentation in the training procession, which avoided overfitting. To evaluate the accuracy of carotid vessel wall segmentation, the sensitivity, specificity and IoU were used.
Results
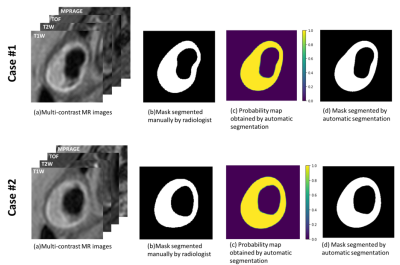
Figure 2 shows the segmented results of carotid vessel wall in multi-contrast images. Probability map was generated at the pixel level. The probabilities of vessel wall area are almost 1.0 and background area are almost 0.0. Figure 3 shows the quantitative evaluation of models trained with sensitivity, specificity and IoU. All evaluation values are larger than 0.75.Discussion
In this work, we proposed an automatic segmentation algorithm of carotid vessel wall in multi-contrast blackblood images based on a fine-tuning U-net neural network. Through copying first convolution layer four times and summing them together, the U-net model could be utilized on the multi-contrast MR images. The concatenate layer between encoder path and decoder path made high resolution of output images.Conclusion
In this work, an automatic segmentation algorithm of carotid vessel wall in multi-contrast blackblood images has been implemented. Training and evaluation on a large-scale clinical data set shows good segmentation performance. In future implementations, we will train a model for each contrast MR images and ensemble them to get a more robust model.Acknowledgements
No acknowledgement found.References
1. Yuan, C., et al., In Vivo Accuracy of Multispectral Magnetic Resonance Imaging for Identifying Lipid-Rich Necrotic Cores and Intraplaque Hemorrhage in Advanced Human Carotid Plaques. Circulation, 2001. 104(17): p. 2051-2056.
2. D Xu, WS Kerwin, T Saam, M Ferguson, and C Yuan. Cascade: Computer aided system for cardiovascular disease evaluation. In Proc ISMRM, page 1922, 2004.
3. Ronneberger O., Fischer P., Brox T. (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J., Wells W., Frangi A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham
4. Long, Jonathan, Evan Shelhamer, and Trevor Darrell. "Fully convolutional networks for semantic segmentation." Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015.
Figures