1020
Optimized 3D Stack-of-Spirals MR Fingerprinting with Hybrid Sliding-Window and GRAPPA Reconstruction1Department of Biomedical Engineering, Zhejiang University, Hangzhou, China, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States
Synopsis
We demonstrate 3D stack-of-spiral MRF acquisition with hybrid sliding-window and GRAPPA (SW+GRAPPA) reconstruction using a constant density spiral (CDS) rather than standard variable density spiral (VDS). The CDS was found to mitigates high-frequency artifacts after in-plane sliding-window (SW) combination and improve the subsequent GRAPPA and dictionary matching reconstruction. The proposed 3D constant density stack-of-spiral MRF allows whole-brain (240×240×192 mm3) parametric mapping at 1 mm isotropic resolution with high quality in 8 minutes.
Introduction
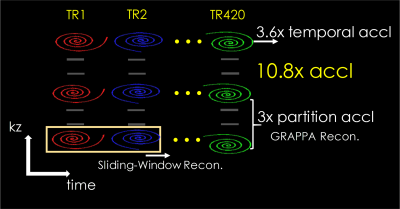
MRF1 has shown great potential for efficient multi-parameter mapping. 3D-MRF acquisitions2-5 enjoy an SNR efficiency benefit over their 2D counterparts, and could help achieve high SNR at high resolutions. However, high resolution imaging with whole-brain coverage can lead to lengthy scans which, in turn, increases its sensitivity to motion. To accelerate 3D-MRF, our recent work 6 combines VDS acquisition with hybrid sliding-window 7 and GRAPPA reconstruction (Fig.1). This enables >10x acceleration through i) 3-fold acceleration in kz encoding and ii) 3.6-fold reduction in the number of TRs for pattern matching (420 instead of 1500 TRs as in [5]). However, 3D-MRF with in-plane resolution of 1 mm or higher exhibits some residual ringing artifacts. In this work, we demonstrate that the use of CDS in place of VDS can mitigate this artifact to achieve high-quality results that outperformed state-of-the-art 3D-MRF method. Interestingly, while it improved the sliding-window reconstruction, the CDS lead to poorer results for standard reconstruction. This finding highlights the need in joint acquisition and reconstruction design/optimization in MRF.Methods
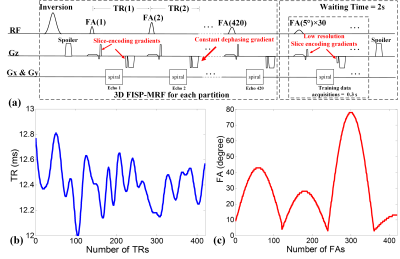
Sequence: 3D FISP sequence8 was implemented for MRF. Figure 2(a) shows the diagram of this partition-by-partition sampled sequence that also incorporates a low-flip-angle training data acquisition into the wait period. The total acquisition time for each partition is 7 seconds for a 420 time-points (tps) acquisition. The TR and FA trains are shown in Fig.2(b&c). CDS k-space trajectory, which consisted of 30 interleaves with zero-moment nulling, was utilized for acquisition. Interleaves were rotated by 12° for each TR with 1400 points per interleaf (7ms readout). To compare the proposed CDS with VDS method, the zero-moment compensated VDS with the same 30 interleaves was used in this study.
SW+GRAPPA reconstruction: SW and NUFFT9 are used to remove in-plane aliasing and create a Cartesian dataset that is fully sampled in-plane. This then allows for a direct application of Cartesian GRAPPA reconstruction to overcome Rz acceleration10. More details can be found in [6].
Data acquisition: Two datasets were acquired on a healthy subject, both with VDS and CDS, using a Siemens Prisma 3T scanner with a 64-channel coil.
i) Accelerated 1 mm isotropic whole-brain data at Rz=3 and 420 time points per partition. Acquisition was performed sagittally, with FOV of 240×240×192 mm3 and Tacq = 8 minutes.
ii) Fully sampled 1×1×4 mm3 whole-brain data, acquired transversally with 48 fully sampled partitions and 1200 time points per partition. Tacq = 15.2 minutes. This data was used to characterize the performance of CDS and VDS at various accelerations. Reconstructions through both standard matching and SW+GRAPPA were performed and compare in each case.
Results
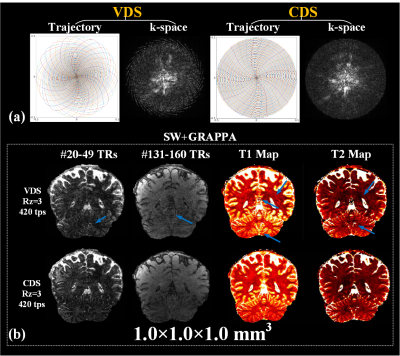
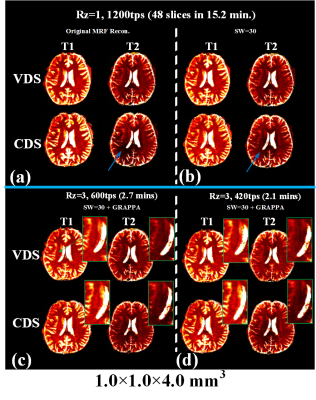
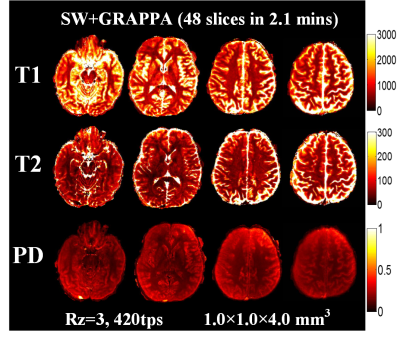
Fig.3a shows a comparison of fully sampled CDS and VDS k-space data at 1mm isotropic obtained via SW across 30 TRs. Significant portion of high k-space data is missing in VDS, which causes ringing/high-frequency artifacts, both in SW images and the resulting T1 and T2 maps as shown in Fig.3b(top-row). These artifacts are effectively removed through CDS as shown Fig.3b(bottom-row), which visibly improved the quality of our highly accelerated 8-minutes 1mm3 whole-brain MRF. Fig.4 shows comparison of results from VDS and CDS at different accelerations, with different reconstructions. For fully sampled data, standard dictionary matching achieves good results for VDS, but poor reconstruction for CDS (Fig.4a). With SW reconstruction, both VDS and CDS achieve high quality results (Fig.4b). Reconstructions at Rz=3 with 600 and 420 time-points are also shown in Fig.4c&d. Here, ringing artifacts, which increased at 420tps, are observed for VDS. On the other hand, CDS achieves good reconstruction even at 420tps. Fig.5 displays high-quality quantitative maps of four representative slices from CDS acquisition at Rz=3 and 420tps, which provide 1x1x4mm3 whole-brain mapping in 2.1 minutes.Discussion
For 3D-MRF with SW+GRAPPA reconstruction, SW plays a key role for high accelerations by reducing the number of necessary time-points and enabling the generation of Cartesian k-space data that is fully sampled in-plane, which facilitates Rz acceleration through GRAPPA. For the original MRF method, VDS performs better than CDS since it oversamples the center of k-space and captures the low frequency information, which is beneficial for “seeing through” highly under-sampled data during the dictionary matching process. However, for highly accelerated 3D-MRF with limited time-points, the removal of in-plane aliasing from VDS through SW and dictionary matching proves insufficient. In this situation, CDS with denser high-frequency k-space coverage effectively suppresses the ringing after SW operation to improve the subsequent GRAPPA and matching reconstruction. This highlights the importance of a joint acquisition and reconstruction design approach for MRF.Acknowledgements
This work was supported in part by NIH research grants: R01EB020613, R01EB019437, R24MH106096, P41EB015896, and the shared instrumentation grants: S10RR023401, S10RR019307, S10RR019254, S10RR023043References
1. Ma D et al., Nature 2013; 495, 187-192.
2. Ma D et al., ISMRM 2016, p.3180.
3. Ma D et al., Magn. Reson. Med. 2016; 75, 2303-2314.
4. Buonincontri G et al., Magn. Reson. Med. 2016; 76, 1127-1135.
5. Ma D et al., Magn. Reson. Med. 2017; doi: 10.1002/mrm.26886.
6. Liao C et al., Neuroimage. 2017; 162, 13-22.
7. Cao X et al., Magn. Reson. Med. 2017; 78, 1579-1588.
8. Jiang Y et al., Magn. Reson. Med. 2015; 74(6), 1621-1631.
9. Fessler JA, J. Magn. Reson. 2007; 188, 191-195.
10. Griswold MA et al., Magn. Reson. Med. 2002; 47, 1202-1210.
Figures