0999
A Fully Convolutional Neural Network for 3D Volumetric Liver Lesion Segmentation1Arterys, San Francisco, CA, United States, 2Radiology, UC San Diego Health, La Jolla, CA, United States
Synopsis
We present an automated approach to liver lesion segmentation in abdominal MRI scans. We use a 3D fully convolutional neural network to segment liver lesions; segmentations are then used to estimate longest linear diameter (LLD) and volume. We show that the median LLD error is 2.01 mm and that these estimates are within limits of clinically usability as part of a semi or fully-automated workflow. Automating lesion segmentation may pave the way for tracking lesion volume and tumor burden as well as treatment response.
Introduction
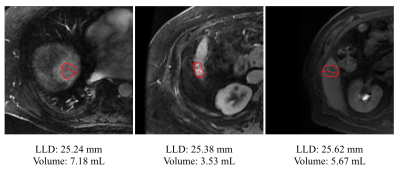
The current clinical standard of care for liver lesions relies on measurements of the longest linear diameter (LLD) to quantify lesion size and measure growth over time1. However, LLD may be a misleading way to measure anisotropic and irregularly shaped lesions2. Volumetric measurements are preferred, as they give a better impression of the overall lesion size (Figure 1). Generating those measurements, however, is time-consuming.
Convolutional neural networks have been successfully used to segment liver lesions in abdominal CT, but very few studies have explored their application to segmenting liver lesions in abdominal MRI3. In this work, we propose a deep-learning-based method for volumetric segmentation of lesions in abdominal MRI. We show that the accuracy of LLD calculated from the segmentations is viable for clinical usage under current guidelines.
Methods
Training Data
For training the model, we use 1,001 annotated lesions from 483 unique abdominal MRI studies and 301 unique patients. DICOM images and lesion locations were collected as part of standard clinical care and contours for the identified lesions were collected using annotators trained in liver lesion segmentation.
Model Architecture
We use a fully-convolutional neural network that operates on 3D patches, centered on the lesion of interest, from multiphasic contrast-enhanced MRI series. The network is a variant of the ENet segmentation architecture4. Training and inference are performed using the Keras5 deep learning package with Tensorflow as the backend.
Evaluation
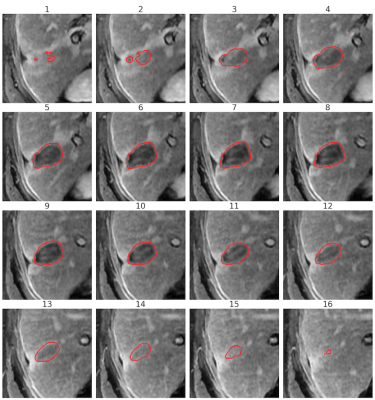
We evaluate our model on a held-out test set of 191 lesions from 83 unique studies and 72 unique patients. Radiologists provided 3D ground truth segmentations for each lesion following the LI-RADS guidelines1. Ground truth LLD and volume were calculated using these contours.
Results
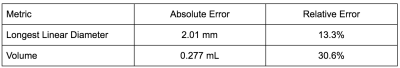
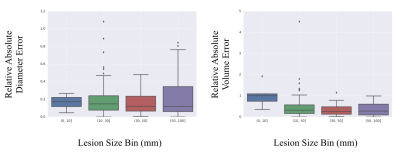
We compare our error averaged across all lesions in the test set. Table 1 summarizes our results - our automated contouring method achieved a median LLD error of 2.01 mm, and a median volume error of 0.277 mL (median lesion size was 15.1 mm and 1.362 mL). Figure 2 presents boxplots for the relative error of the two metrics of interest, binned by lesion size, and demonstrates that our model performance is similar across lesions of different sizes.Discussion
Median absolute LLD error from the CNN was significantly below the LI-RADS “threshold growth” threshold, suggesting that our automated contouring method could potentially be used as a standalone, fully-automated tool for determining LLD. Future work may focus on assessing the reliability of volumetric segmentations to aid in clinical management.Conclusion
We present an automated approach to lesion segmentation in abdominal MRI. We use a 3D fully convolutional neural network to segment liver lesions and estimate longest linear diameter (LLD) and volume. We show that the median LLD error is 2.01 mm and that these estimates may be used as part of a semi-automated clinical workflow. Additionally, we demonstrate that volumetric estimates are feasible from MRI and can enable more precise tracking of tumor burden over time.Acknowledgements
No acknowledgement found.References
- American College of Radiology. CT/MR Liver Imaging Reporting and Data System version 2017. Accessed October 2017, from http://www.acr.org/Quality-Safety/Resources/LIRADS.
- Bernardin, Livia & O'Flynn, Elizabeth & M Desouza, Nandita. (2013). Functional imaging biomarkers for assessing response to treatment in liver and lung metastases. Cancer imaging : the official publication of the International Cancer Imaging Society. 13. 482-494. 10.1102/1470-7330.2013.0047.
- Christ, P. et al (2017, February 23). Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks. https://arxiv.org/abs/1702.05970
- Paszke, A., Chaurasia, A., Kim, S., & Culurciello E. (2016, June 7). ENet: A Deep Neural Network Architecture for Real-Time Semantic Segmentation. https://arxiv.org/abs/1606.02147
- Chollet, F., Keras, (2017), GitHub repository, https://github.com/fchollet/keras
Figures