0958
Whole-Tumor Histogram and Texture Analyses of Diffusion Tensor Imaging for Evaluation of IDH1 Mutation and 1p/19q Codeletion Status in WHO grade II gliomas1Radiology, Ewha Womans University College of Medicine, Seoul, Republic of Korea, 2Yonsei University College of Medicine, Seoul, Republic of Korea
Synopsis
We analyzed the histogram and texture features of apparent diffusion coefficient (ADC) and fractional anisotropy (FA) based on entire tumor to determine isocitrate dehydrogenase 1 (IDH1) mutation and 1p/19q codeletion status in WHO grade II gliomas. Regions of interest were drawn on ADC and FA maps of 93 grade II gliomas. Histogram and texture analyses were performed. The areas under the curve (AUC) for IDH1-wildtype prediction was 0.853, and AUC for 1p/19q codeletion prediction was 0.807. The whole-tumor histogram and texture features of ADC and FA maps are useful in predicting the molecular status in WHO grade II gliomas.
INTRODUCTION
According to the 2016 the World Health Organization (WHO) classification, the molecular subtypes of WHO grade II gliomas are divided into three classes: isocitrate dehydrogenase (IDH)-wildtype, IDH-mutant and no 1p/19q codeletion, and IDH-mutant and 1p/19q-codeleted.1 Different molecular subtypes have been reported to have prognostic differences and different chemosensitivity. 2,3 IDH gene mutation and 1p/19-codeletion may reflect alterations in tumor cell proliferation and microvessel density, which may manifest characteristic features on apparent diffusion coefficient (ADC) and fractional anisotropy (FA) parameters. The texture analysis features shows the characteristics of the entire tumor and has the advantages of noninvasively quantifying tumor features such as tumor uniformity, heterogeneity, smoothness, randomness, and symmetry.4 Thus, the purpose of this study was to characterize the histogram and texture analyses of ADC and FA maps based on entire tumor volume data to determine IDH1 mutation and 1p/19q codeletion status in WHO grade II gliomas.
METHODS
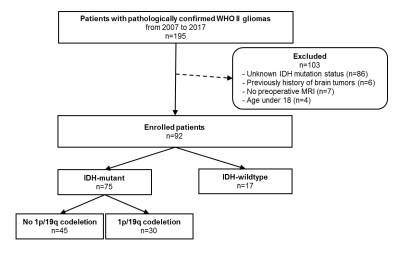
Ninety-three WHO grade II gliomas patients with known IDH1 mutation and 1p/19q
codeletion status (18 IDH1-wildtype, 45 IDH1-mutant and no 1p/19q codeletion,
30 IDH1-mutant and 1p/19q codeleted tumors) underwent diffusion tensor imaging.
Regions of interest were drawn on every section of the ADC and the FA map
containing the tumor and were summated to derive volume-based data of the
entire tumor. Histogram and texture analyses were correlated with the IDH1
mutation and 1p/19q codeletion status. The predictive powers of imaging
features for IDH1-wildtype tumors and 1p/19q codeletion status in IDH1-mutant
subgroups were evaluated using the least absolute shrinkage and selection
operator (LASSO).
RESULTS
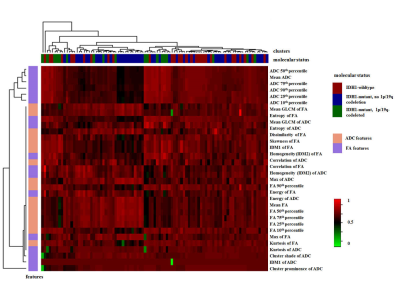
Various histogram and texture parameters were significantly different according to IDH1 mutation and 1p/19q codeletion status. The skewness and energy of ADC, FA 10th percentile, 25th percentile, and correlation of FA were independent predictors of an IDH1-wildtype in LASSO. The areas under the curve (AUC) for the prediction model were 0.853. The skewness and cluster shade of ADC, energy and correlation of FA were independent predictors of an 1p/19q codeletion in IDH1-mutant tumor in LASSO. The AUC was 0.807.DISCUSSION
Preoperative prediction of the molecular classification of WHO grade II gliomas is useful to guide the treatment decision and predict the prognosis. WHO grade II gliomas are heterogeneous on both genetic and histopathological levels with intratumoral spatial variation 5; in the present study, these were evaluated using an ADC and FA histograms and texture analyses based on the whole tumors.
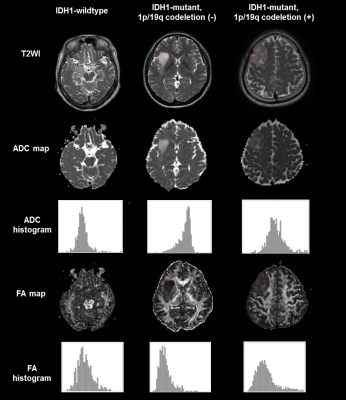
In WHO grade II gliomas, IDH1 wildtype tumors showed lower ADC values and higher FA values than IDH1 mutant tumors. IDH mutation is associated with lower tumor cellularity 6 and it explains why the presence of an IDH mutation is a favorable prognostic marker in glioma.7 The lower FAs in WHO grade II gliomas with IDH mutations may be due to the lower rate of proliferation and aggressiveness, as well as the lower tumor cell densities. The IDH1 wildtype group had higher skewness and energy of ADC, 10th and 25th percentile of FA, whereas the IDH1 mutant group demonstrated higher FA correlation. Thus, IDH1 wildtype group demonstrated higher ADC orderliness, whereas the IDH1 mutant group had a higher frequency of similar FA value regions. These results may be due to different histopathologies which are imperceptible to visual analyses.
Low-grade gliomas bearing an IDH mutation with a 1p/19q codeletion are associated to have a favorable prognosis and to be sensitive to chemotherapy with alkylating agents. 2,8,9 In our study, IDH mutant with 1p/19q-codeletion tumors showed lower ADC and higher FA values than IDH mutant without 1p/19q-codeletion tumors. IDH mutant with 1p/19q-codeletion tumors are markedly infiltrated by perineuronal satellitosis with more persistent neurons, which may explain their higher FA values.10 The sparing of neurons with lesser volume of invasion may lead to less edema, which may explain the lower ADC values in such tumors.10 Also, the micro-calcifications which are seen in up to 90% of IDH mutant with 1p/19q codeletion tumors 10 may also contribute to lower ADC values due to the lack of water movement. The 1p/19q codeletion group demonstrated higher ADC skewness, whereas the no 1p/19q codeletion group had higher FA energy and FA correlation. Thus, the 1p/19q codeletion group showed more ADC asymmetry, whereas the no 1p/19q codeletion group had higher FA organization and more regions with similar FA values.
CONCLUSION
Whole-tumor histogram and texture features of the ADC and FA maps are useful in predicting the IDH1 mutation and 1p/19q codeletion status in WHO grade II gliomas.Acknowledgements
The authors report no conflict of interest concerning the materials or methods used in this study or the findings specified in this paper.References
1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta neuropathologica 2016;131:803-820.
2. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. New England Journal of Medicine 2015;372:2481-2498.
3. Metellus P, Coulibaly B, Colin C, et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta neuropathologica 2010;120:719-729.
4. Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta neuropathologica 2010;120:567-584.
5. Paulus W, Peiffer J. Intratumoral histologic heterogeneity of gliomas. A quantitative study. Cancer 1989;64:442-47
6. Bralten LB, Kloosterhof NK, Balvers R, et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Annals of neurology 2011;69:455-463.
7. Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. Journal of Clinical Oncology 2009;27:4150-4154.
8. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. New England Journal of Medicine 2015;372:2499-508
9. van den Bent MJ. Practice changing mature results of RTOG study 9802: another positive PCV trial makes adjuvant chemotherapy part of standard of care in low-grade glioma. Neuro-oncology 2014;16:1570-74
10. Khayal IS, McKnight TR, McGue C, et al. Apparent diffusion coefficient and fractional anisotropy of newly diagnosed grade II gliomas. NMR in Biomedicine 2009;22:449-455.
Figures