0912
Down-regulation of phospholipid biosynthesis is a unique metabolic feature of mutant IDH1 gliomas mediated by autophagy of the endoplasmic reticulum1Radiology, University of California San Francisco, San Francisco, CA, United States, 2Neurosurgery, University of California San Francisco, San Francisco, CA, United States
Synopsis
Virtually every cancer studied so far shows elevated choline and ethanolamine phospholipid metabolism, which has emerged as a metabolic hallmark of cancer. Here, we show that, unusually, low-grade gliomas carrying a mutation in isocitrate dehydrogenase 1 (IDHmut) down-regulate phosphatidylcholine and phosphatidylethanolamine biosynthesis and steady-state levels. Mechanistically, this down-regulation is mediated via autophagic degradation of the endoplasmic reticulum, the site of phospholipid biosynthesis. Importantly, the autophagy inhibitor chloroquine restores phospholipid levels and abrogates IDHmut tumor growth, identifying a potential therapeutic opportunity. Thus, our study demonstrates that IDHmut gliomas uniquely down-regulate phospholipid biosynthesis and that this phenomenon can be exploited for therapy.
Introduction
Elevated levels of phosphocholine (PC) and phosphoethanolamine (PE) are MR-detectable metabolic hallmarks of cancer1. Unusually, low-grade gliomas carrying a mutation in the metabolic enzyme isocitrate dehydrogenase 1 (IDHmut) down-regulate PC and PE relative to wild-type IDH1 (IDHwt) gliomas2,3. PC and PE are precursors of the membrane phospholipids phosphatidylcholine (PtdCho) and phosphatidylethanolamine (PtdE)4. Therefore, the goal of this study was to investigate whether PtdCho and PtdE levels are also down-regulated in IDHmut gliomas.Methods
We performed our experiments on two models (immortalized normal human astrocyte (NHA) and U87 glioblastoma-based models) genetically-engineered to express wild-type (U87IDHwt/NHAIDHwt) or mutant IDH1 enzyme (U87IDHmut/NHAIDHmut)3. MR studies of cell extracts were performed on a 500MHz Bruker spectrometer. Metabolites were extracted by dual-phase extraction3 and the lipid fraction resuspended in a 2:1 mixture of deuterated chloroform and deuterated methanol containing 60mM CsEDTA. Steady-state phospholipid levels were determined from proton-decoupled 31P-MR spectra (30°flip angle, 2.6s relaxation delay, 1440 scans). To measure de novo phospholipid biosynthesis, cells were labeled with 56μM [1,2-13C]-choline and [1,2-13C]-ethanolamine, and proton-decoupled 13C-MR spectra (30°flip angle, 3s relaxation delay, 2048 acquisitions) acquired. Peak integrals were quantified (Mnova), corrected for saturation and normalized to cell number and external reference (trimethylsilylpropionate for 13C-MR and methylene diphosphonic acid for 31P-MR). Kinetic build-up of 13C-Ptdcho and 13C-PtdE synthesis was analyzed by non-linear regression (GraphPad Prism). Autophagy was inhibited using 50μM chloroquine (CQ) for 16h. Autophagic flux was quantified by measuring LC3-II levels in the presence of the lysosomal inhibitor bafilomycin A1 by immunoblotting as recommended5. CCT activity was determined as described6. For ECT activity, cells were lysed (50mM HEPES pH 7, 5mM EDTA) and combined with reaction mix (50mM Tris-HCl pH 8, 5mM DTT, 10mM cytidine triphosphate, 5mM PE, 25mM MgCl2), followed by acquisition of proton-decoupled 31P-MR spectra as above. ECT activity was calculated by linear regression of the kinetics of CDP-ethanolamine production. STORM imaging of calnexin fluorescence7, immunoblotting8 and clonogenicity8 were performed as described. U87IDHmut orthotopic tumor xenografts were generated by intracranial implantation of U87IDHmut cells in nude mice9. MR imaging was performed on a 14.1T vertical MR system (Agilent). Once the tumor reached 2-3mm, this time point was considered day zero and mice treated intraperitoneally with saline or 60mg/kg CQ. Experiments were repeated five times and significance assessed using an unpaired Student’s t-test assuming unequal variance with p<0.05 considered significant (* = p<0.05; ** = p<0.01; *** = p<0.005).Results
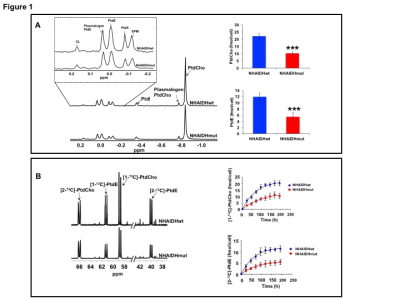
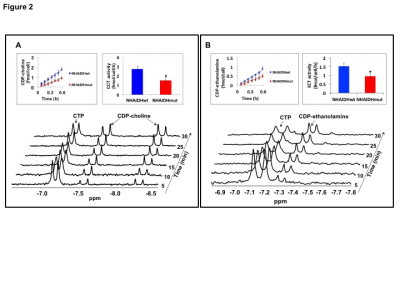
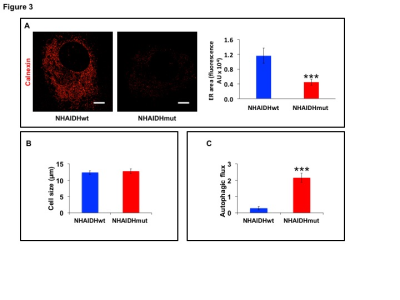
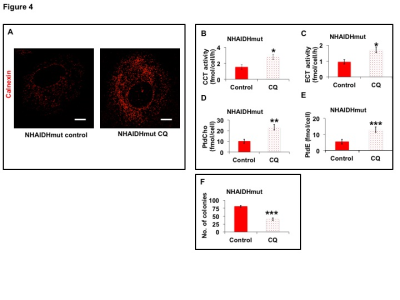
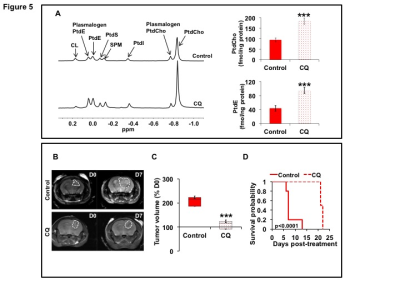
31P-MRS (Fig.1A) of the lipid fraction of cell extracts showed a significant reduction in steady-state PtdCho and PtdE in NHAIDHmut relative to NHAIDHwt. 13C-MRS (Fig.1B) of cells labeled with [1,2-13C]-choline and [1,2-13C]-ethanolamine for different time points showed a significant reduction in de novo synthesis of 13C-PtdCho and 13C-PtdE in NHAIDHmut relative to NHAIDHwt. Next, using 31P-MR-based assays, we examined the activities of CCT and ECT, the rate-limiting enzymes for PtdCho and PtdE biosynthesis4. We found a significant reduction in CCT (Fig.2A) and ECT (Fig.2B) activity in NHAIDHmut relative to NHAIDHwt. CCT and ECT are localized to the endoplasmic reticulum (ER), which is the principal site for phospholipid biosynthesis4. We therefore measured ER area using super-resolution stochastic optical resonance microscopic (STORM) imaging7 and found a significant reduction in NHAIDHmut relative to NHAIDHwt (Fig.3A), while cell size remained unchanged (Fig.3B). Autophagy of the ER (“ER-phagy”) can regulate ER area10 and so we questioned whether ER-phagy was activated in IDHmut cells. We found that autophagic flux was higher in NHAIDHmut relative to NHAIDHwt (Fig.3C). Importantly, we obtained similar results with the U87 model (data not shown). To further confirm our findings, we inhibited autophagy using CQ. We found that CQ restored ER area (Fig.4A), CCT activity, (Fig.4B) ECT activity (Fig.4C) and phospholipid levels (Fig.4D-4E) confirming the link between ER-phagy and phospholipid biosynthesis. Furthermore, CQ significantly inhibited colony formation (Fig.4F), indicating that IDHmut cells in which autophagy is inhibited would be unable to form tumors, pointing to an important therapeutic opportunity. Finally, to confirm the significance of our findings in vivo, we examined the effect of CQ on orthotopic U87IDHmut tumor xenografts. CQ increased phospholipid levels (Fig.5A), significantly reduced tumor growth (Fig.5B-5C) and prolonged survival (Fig.5D) of mice bearing U87IDHmut orthotopic tumor xenografts.Conclusions
Cancer cells normally up-regulate phospholipid biosynthesis to meet the demands of uncontrolled cell proliferation. Unusually, our MR findings and associated studies indicate that IDHmut glioma cells down-regulate phosphatidylcholine and phosphatidylethanolamine biosynthesis as a result of autophagic degradation of the endoplasmic reticulum. Reduced phospholipid biosynthesis therefore constitutes a unique metabolic biomarker of IDHmut gliomas. Importantly, our studies underscore the therapeutic potential of targeting this metabolic reprogramming for treatment of IDHmut gliomas.Acknowledgements
NIH R01CA172845, NIH R01CA197254, NIH R01CA154915, NIH P41EB013598, UCSF Brain Tumor Loglio Collective and NICO.References
1) Cheng et al., Front Oncol, 6:266, 2016. 2) Esmaeili et al., Cancer Res, 74:4898-4907, 2014. 3) Izquierdo-Garcia et al., PLOS ONE, 10:e0118781, 2015. 4) Gibbelini & Smith, IUBMB Life, 62(6):414-28, 2010. 5) Klionsky et al., Autophagy, 12 (1): 1-222, 2016. 6) Ward et al., PLOS ONE, 8:e62610, 2013. 7) Huang et al., Science, 319 (5864):810-813, 2008. 8) Izquierdo-Garcia, Viswanath et al., Cancer Res, 75 (15): 2999-3009, 2015. 9) Chaumeil et al. Nat Comm, 4: 2429-2441, 2013. 10) Khaminets et al., Nature, 522 (7556): 354-8, 2015.Figures