0902
Improved Prediction of the Final Infarct from Acute Stroke Neuroimaging Using Deep Learning1Tsinghua University, Beijing, China, 2Stanford University, Stanford, CA, United States
Synopsis
Magnetic Resonance Imaging (MRI) is a widely-used technique for clinics. Its advantages in providing multiple complimentary contrasts make it the best image tool for detecting presenting lesions in the brain. A lot methods have been proposed for lesion detection and segmentations using machine learning techniques. It is more sophisticated than common computer vision tasks since the estimation of treatment outcomes are not merely determined by lesions captured by current MR images. We targeted to develop an algorithm, based on 3D Convolutional Neural Network, to predict the final lesion shown on day-90 scans by processing the day-0 acute stroke images.
Introduction
Predicting the final infarct of an acute ischemic stroke using magnetic resonance imaging (MRI) is valuable for triage and prognosis assessment[1]. We developed and tested a deep learning model to predict the final stroke lesion from acute stroke multi-contrast MR images, using a 3D Convolutional Neural Network (CNN) architecture with improved designs for multi-scale and patch-based learning.Methods
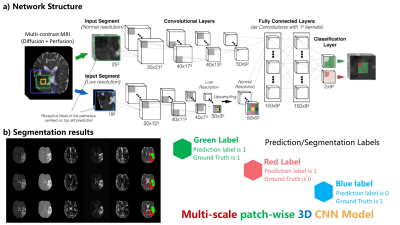
The detailed network structure of our model is shown in Figure 1 below.
- Model structure
Compared with traditional CNN networks, our proposed model has three main contributions and features:
* (1) A three-dimensional CNN[2,3] was implemented which utilizes available spatial information more effectively and exploits relationships between different slices.
* (2) Our patch-wise approach uses several small 3D patches taken from the original images/volumes as inputs. This approach focuses on local voxel information, minimizes the impact of distant unrelated voxels, and handles image/volumes with different dimensions without explicitly cropping into a standard dimension. Training on patches also prevents over-fitting by extracting from each image thousands of samples.
* (3) The multi-scale structure learns the lesion prediction from patches with different resolutions to fuse information at various scales. Segmentation based on a single scale image cannot fully capture both local information and global contextual information simultaneously. Therefore, we use two stacks of patches with different scales to extract both local and global contextual information.
- Validation
Our proposed 3D CNN model was trained and tested using the MICCAI ISLES 2017 challenge dataset. This dataset consists of 43 cases with acute DWI-ADC and PWI maps registered to the annotated final infarct segmentation at day-90th. We split labeled data into two sets: 77% for training and 23% for testing. Similar to the criteria of the challenge, the Dice Score Coefficient (DSC) was used as the major quality metric to evaluate the model’s segmentation performance.
Results
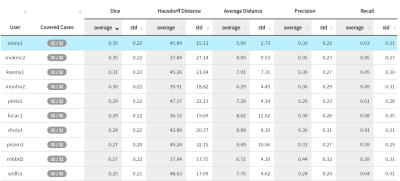
With proper data augmentation using rigid transformations, we achieved a mean DSC of $$$ 0.35 \pm 0.20 $$$ on the testing set. Our model performs better on test dataset samples with large stroke lesions, reaching a DSC around 0.70. As a comparison, the winning entry for the ISLES 2016 challenge achieved a DSC of $$$ 0.31\pm 0.24 $$$. Our proposed method also outperforms the results in previous research (threshold-based, cluster-based, generalized linear model, etc.), and is the top entry in the ISLES 2017 public leaderboard as shown in Figure 2.Discussion
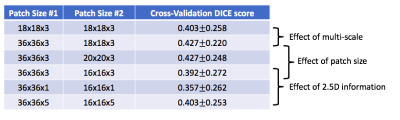
We compared the results for the model using single or multiple scales and with different patch sizes. From Figure 3, we showed that the implemented multi-scale and 3D patch-wise approach with specific patch size is achieving a good balance of model complexity and performance.
Given the small datasets, our detailed analysis revealed worse performance in the few cases with tiny lesions, which shall be further improved with more tuning. To improve the segmentation results and to ensure that the proposed method can generalize well (including rare pathologies), we will explore more architecture options and additional methods, such as using a Conditional Random Field (CRF) [4] and a Generative Adversarial Network (GAN)[5].
Conclusion
We demonstrated that a deep learning approach with an advanced network architecture can predict the final stroke lesion accurately based on acute diffusion and perfusion MRI. Given its inherent speed, high performance, and capacity for further improvement, deep learning is a promising method for MRI based stroke outcome prediction.Acknowledgements
No acknowledgement found.References
1. Rekik, Islem, et. al. NeuroImage: Clinical 1.1 (2012): 164-178.
2. Kamnitsas, Konstantinos et al. Medical image analysis. 36, 61–78, (2017)
3. Ji S, et al. IEEE transactions on pattern analysis and machine intelligence, 2013
4. Krhenbhl P, Koltun V. Advances in neural information processing systems. 2011: 109-117.
5. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Advances in neural information processing systems. 2014: 2672-2680.
Figures