0893
Classification of Dense Tumor, Tumor Necrosis and Tumor Infiltration in Glioma: Machine Learning and Diffusion MRIZezhong Ye1, Xiran Liu2, Joshua Lin1, Liang Wang2, Richard Price3, Peng Sun1, Jeff Viox1, Sonika Dahiya4,5, Albert Kim3, Jr-Shin Li2, and Sheng-Kwei Song1
1Radiology, Washington University School of Medicine, St. Louis, MO, United States, 2Electrical & System Engineering, Washington University in St. Louis, St. Louis, MO, United States, 3Neurological Surgery, Washington University School of Medicine, St. Louis, MO, United States, 4Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, United States, 5Immunology and Pathology, Washington University School of Medicine, St. Louis, MO, United States
Synopsis
Here we introduce a diffusion MR-based imaging technique - Diffusion MRI Histology (D-Histo), to detect and differentiate various co-existing tumor pathologies including high-cellularity tumor (tumor), tumor necrosis (necrosis) and tumor infiltration (infiltration) within high grade glioma. We incorporated a support vector machine algorithm to generate an automation framework to predict locations of tumor lesion, necrosis and infiltration. The mean predictive accuracy of the D-Histo SVM classifier for tumor lesion, necrosis and infiltration were 91.9%, 93.7% and 87.8%. DTI-based prediction under the same framework resulted in 44.4%, 56.0% and 43.0% accuracy for the three pathologies.
Introduction
High-grade glioma often presents complicated tumor micro-environment containing co-existing tumor pathologies, which includes high tumor cellularity, central necrosis, micro-vascular proliferation and tumor infiltration. These pathological hallmarks set Grade IV GBM from Grade III glioma such as anaplastic astrocytoma, where the latter commonly associates with increasing tumor cellularity alone. Restricted diffusion caused by increased cellularity in glioma could be detected by DWI-derived ADC to classify glioma;1 except for between Grades III and IV. The shared characteristic of increased cellularity between the two higher-grade gliomas requires additional spectrum for accurate diagnosis. Here we applied a novel MRI-based imaging technique called diffusion MRI histology (D-Histo), and support vector machine (SVM) to detect and classify different pathologies in glioma.Materials and Methods
Brain tumor specimen: 14 high-grade glioma patients were recruited (Table 1) for the study. 22 brain tumor specimens were procured and fixed in 10% formalin and transferred to PBS. MRI and histological staining: Brain tumor specimens were examined using a 4.7T MR Varian scanner and a home-made surface coil. A spin-echo diffusion-weighted sequence with 99 diffusion-encoding directions with maximum b-values of 3000 s/mm2 was employed to acquire DWI (in-plane resolution 0.25x0.25x0.5 mm2). The specimens underwent sequential sectioning at 5 μm thickness and was stained with H&E. Data analysis: An in-house software analyzed D-Histo and DTI data. H&E images were reviewed by an experienced neuropathologist. D-Histo maps were linearly co-registered with corresponding H&E images, allowing MRI voxels to be labeled by the histological gold standard. Machine learning: A supervised machine-learning-framework adopting SVM with RBF-kernel algorithm was used. D-Histo-derived restricted, hindered, and anisotropic fractions were used as the feature values for each voxel. We evaluated classification accuracy by pooling voxels from different classes and within each class. Receiver operating characteristics (ROC) analyses were performed to assess how well one tumor pathology could be distinguished from the rest.Results
Figure1 displayed a representative case of a GBM specimen (Fig. 1A). T2WI, DWI and ADC map (Fig. 1B,C,D) displayed unclear or false-positive signals for dense tumor (red arrows). Hyper-intensities in D-Histo highly-restricted-fraction (Fig. 1G), restricted-fraction (Fig. 1H) and hindered-fraction (Fig. 1I) maps correspond well with tumor infiltration, dense tumor and tumor necrosis indicated by H&E and GFAP (Fig. J,K). 5220 voxels from 18 glioma specimens were distributed equally to training and validation groups to construct a robust SVM classifier. Training and validation accuracies were 93.8% and 93.9%, respectively. The 4 remaining specimens were set as testing group. D-Histo-SVM prediction (Fig. 2B) agreed highly (overall-accuracy of 95.6%) with histology classification (Fig. 2A). Prediction accuracy of the dense tumor, necrosis and tumor infiltration were 97.3%, 100% and 85.1%. Each sample was also tested to evaluate the performance of D-Histo-SVM classifier on a single subject. Representative D-Histo voxels were selected to be displayed with their corresponding H&E images. 83.6% of imaging voxels in B94 (Fig. 3A) were predicted correctly to be tumor infiltration. 98.8% and 100% of voxels in B95 (Fig. 3B) were correctly predicted to be dense tumor and necrosis. Prediction accuracies for dense tumor, necrosis and tumor in B122 (Fig. 3C) were 92.6%, 96.8% and 84.8%. 99.7% of total voxels in B128 (Fig. 3D) were correctly predicted to be dense tumor. To avoid selection bias between the training-validation and prediction groups, 7319 distinct group combinations were run, in which mean values and 95% CI were calculated (Table 2A, all samples). Dense tumor, necrosis and infiltration had average prediction accuracies of 91.7%, 93.7% and 87.8%. Although testing group was excluded when constructing the classifier, it is still possible that some of the testing data would correlate with some of the training data (i.e. they come from the same subject). We remedied this possibility by calculating 260 training-testing combinations, in which subjects in the two groups did not overlap (Table 2B, different subjects). Dense tumor, necrosis and infiltration predictions had 90.8%, 92.0% and 83.2%, accuracies. We performed ROC analyses to test D-Histo-SVM classifier’s ability to differentiate one tumor pathology from the rest (Table 2B). The classifier demonstrated AUCs of 0.978, 0.977 and 0.991 in distinguished between dense tumor and non-dense tumor, between necrosis and non-necrosis, and between infiltration and non-infiltration. Similar results were seen for different subject condition. DTI was less accurate in predicting, with lower AUC values than D-Histo (Table 2B).Discussions and Conclusions
We demonstrated D-Histo could better reflect pathological classification of tumor grades by more accurately characterizing different tumor pathologies that are critical to clinical diagnosis and prognosis. We also differentiated tumor infiltration and tumor necrosis from other tumor pathologies, which could allow better complete tumor removal during surgery and better treatment evaluation.Acknowledgements
This work was supported in part by NIH R01-NS047592, P01-NS059560, U01-EY025500, National Multiple Sclerosis Society (NMSS) RG 4549A4/1, RG-1507-05315.References
1. Yamasaki F, Kurisu K, Satoh K, et al. Apparent diffusion coefficient of human brain tumors at MR imaging. Radiology. Jun 2005;235(3):985-991.Figures
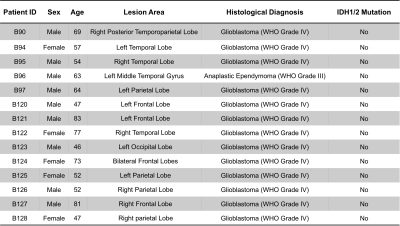
Table 1. Patient information.
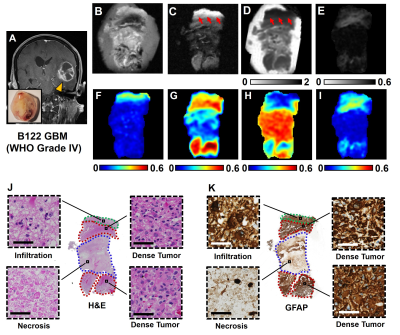
Figure
1. Brain
specimen was resected from a GBM patient (A). T2W image (B), DW images (C) and ADC map (D) showed ambiguous signals
or false-positive information for dense tumor (red arrows). DTI-FA (E) and
D-Histo anisotropic-fraction (F) maps both showed anisotropy in dense tumor
areas. Regions of tumor infiltration (J&K, green outline), dense tumor
(J&K, red outline) and tumor necrosis (J&K, blue outlines) revealed by
the H&E (J) and GFAP (K) images were highly consistent with
hyperintensities in D-Histo highly-restricted-fraction (G), restricted-fraction
(H) and hindered-fraction (I) maps. Magnified views from H&E and GFAP
showed micro-structures of these tumor pathologies.
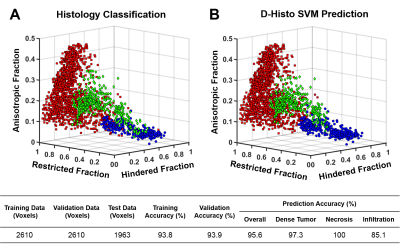
Figure 2. A total of 1963 testing imaging voxels were
divided in to dense tumor (A, red squares), necrosis (A, blue spheres) and
tumor infiltration (A, green diamonds) voxels and color-coded based on their
histological labels. Based on a training (2610 training voxels and 93.8%
training accuracy) and validation (2610 training voxels and 93.9% validation
accuracy) process, D-Histo SVM prediction displayed high agreement with
histology classifications (A) of dense tumor, necrosis and tumor infiltration (B)
with an overall accuracy of 95.6%. Dense tumor, necrosis, and tumor infiltration
were 97.3%, 100% and 85.0% accurate, respectively (C).
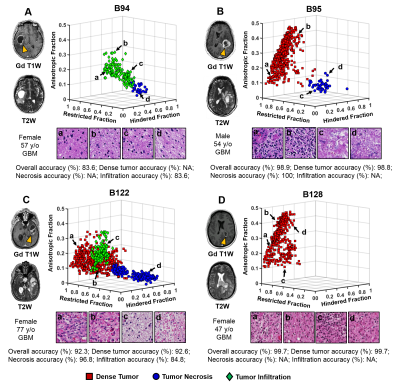
Figure 3. To evaluate the performance of D-Histo
SVM on individual subject, samples from four different subjects were
individually tested. Representative
D-Histo voxels were selected to be displayed with their corresponding H&E
images. In B94, 83.6% of imaging voxels were correctly predicted to be tumor
infiltration (A). 98.8% of dense tumor voxels and 100% of necrosis voxels in
B95 were correctly (B). The correct rates of prediction for dense tumor,
necrosis and tumor infiltration were 92.6%, 96.8% and 84.8% in B122 (C). 99.7%
of total voxels were correctly predicted to be dense tumor in B128.
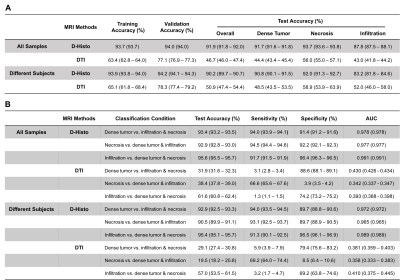
Table 2. Evaluation of D-Histo and DTI SVM
classifiers as indicators of dense tumor, infiltration and necrosis. Values
were summarized as the format of mean (95% Confidence Interval). Section A
shows classification of three different tumor pathologies. Section B shows pairwise
classification and ROC analysis for one tumor pathology versus the rest ones.
All Samples summarize results from 7315 different combinations of training,
validation and test samples. Different Subjects summarized results of 284
combinations that 4 test samples came from different subjects with training and
validation dataset to avoid the internal correlation between test data and
training data.