0877
Modeling Motor Task Activation from Task-free fMRI with Machine Learning: Predictions and Accuracy in Individual SubjectsElizabeth Zakszewski1, Alexander Cohen1, Chen Niu2, Xiao Ling2, Oiwi Parker Jones3, Saad Jbabdi3, Ming Zhang2, Maode Wang2, and Yang Wang1
1Medical College of Wisconsin, Milwaukee, WI, United States, 2First Affiliated Hospital of Xi'An Jiaotong University, Shaanxi Xi'an, China, 3Oxford Centre for Functional Magnetic Resonance Imaging of the Brain (FMRIB), University of Oxford, Oxford, United Kingdom
Synopsis
This study is aimed to apply a newly developed machine learning approach to predict individual motor performance from resting state functional MRI. Our data demonstrate that resting state fMRI even using conventional EPI protocols can predict individual motor performance. Our results suggest that the novel machine learning model could more accurately predict motor function at the individual level, compared to the independent component analysis method.
Introduction
Functional magnetic resonance imaging (fMRI) is often used in surgical planning to map out important areas of the brain related to, for example, motor or language function. However, clinical patients can’t always comply with the tasks required for this type functional mapping. Resting state fMRI (rs-fMRI) creates maps of correlated areas of the brain over time and does not require a task. Traditionally, data-driven independent component analysis (ICA) is applied to rs-fMRI data to create brain network maps analogous to those created with task fMRI.1-3 Most recently, a novel model based on machine learning (ML) has been proposed to accurately predict individual differences in brain activity and highlights a coupling between brain connectivity and function that can be captured at the level of individual subjects.3,4 Here, we compare this novel model with conventional ICA approach, to predict motor task activation maps from rs-fMRI data alone, using the task fMRI activation maps from same subjects as reference.Methods
In total, 28 healthy volunteers were scanned on a 3T MR scanner. For each subject, both structural imaging (3D SPGR 1x1x1 mm3) and fMRI scans (including resting state and task) using the BOLD EPI sequence (TR/TE = 2500/30ms, FA=90, voxel size=3.75x3.75x3mm3, 132 imaging frames) were acquired. In addition to the rs-fMRI acquisition, same subjects also performed a bilateral hand tapping task fMRI. Predictions of motor activation maps were made from rs-fMRI data using newly reported methods of Parker Jones, et al.5 Briefly, all rs-fMRI data was preprocessed using the Human Connectome Project (HCP) minimal preprocessing pipeline6 and converted to the CIFTI format. Features were extracted from the combined rs-fMRI data using dual-step PCA followed by ICA to create a set of 40 group-level feature maps. These were combined with individual rs-fMRI data to create a feature map for each subject. A machine learning algorithm was then “trained” to map from individual feature maps (predictors) to task activation. The beta coefficients were averaged with a “leave-one-out” analysis (leaving out that subject’s beta, creating a model that has not “seen” that subject) for each subject to create a predicted task activation map. A Pearson correlation coefficient was calculated between each subject’s predicted and each other subject’s task map. The matrices were evaluated for diagonality. Gray level thresholds were calculated for each activation map using Otsu’s method and used to binarize the maps so that a Dice overlap coefficient could be calculated between predicted and task maps. Matrices were then row- and column-normalized and comparisons were made between the matrices.Results
Overall, the novel method performed better than the standard ICA method in predicting motor task activation maps. A qualitative comparison (Fig 1, example subject) shows visual similarity in the maps using task activation as reference. Quantitatively, matrices of all subjects compared with all other subjects were created for both the Pearson and Dice values and were row-and column normalized (Fig 2). When the ICA method is used as a predictor, the correlation matrix shows no discernable pattern. However, when the model training method was used to create predictions, a slight diagonal-dominant pattern can be seen for both the correlation and the Dice overlap matrices. This indicates better ability to predict subject-specific maps using the novel method.Discussion and Conclusion
Our results overall showed greater correlation between a predicted map and the task-based map of the same subject than between a predicted map and a task-based map of different subjects. Visual comparison shows good correspondence between maps for a single subject, which means that as a tool for surgeons the ML method matches more closely with actual task activation and potentially superior to the existing ICA method. The ML method also correctly predicts activations in the visual cortex that were not predicted with the ICA. This may be useful in some situations with multi-cortex involvement. The correlation matrices here are not as diagonally dominant as those shown in previous works,4,5 however those works used more data (about 100 subjects each), with one study drawing on high resolution HCP data. Our study has only used 28 subjects with data of more conventional fMRI protocol. In current common clinical settings, a clinical population would look more like our subjects than like the HCP data, so this work is an important preliminary test of the method. The results show promise for a novel ML method to map functional motor areas using conventional rs-fMRI protocols without the need for task compliance. This approach will be very useful in some clinical settings.Acknowledgements
This work was supported by a Daniel M. Soref Charitable Trust Grant (to Y.W.).References
1. Smith SM, Fox PT, Miller KL, et al. Correspondence of the brain's functional architecture during activation and rest. PNAS. 2009; 106(31): 13040–13045. 2. Calhoun VD, Liu J, Adali T. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage. 2009;45(1):S163-S172. 3. Branco P, Sexias D, Deprez S, et al. Resting-State Functional Magnetic Resonance Imaging for Language Preoperative Planning. Front Hum Neurosci. 2016; doi: 10.3389/fnhum.2016.00011 4. Tavor I, Parker Jones O, Mars RB, et al. Task-free MRI predicts individual differences in brain activity during task performance. Science. 2016;352(6282):216-220. 5. Parker-Jones O, Voets NL, Adcock AE, et al. Resting connectivity predicts task activation in pre-surgical patients. Neuroimage – Clinical. 2017;13:378-385. 6. Glasser MF1, Sotiropoulos SN, Wilson JA, et al. WU-Minn HCP Consortium. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105-24.Figures
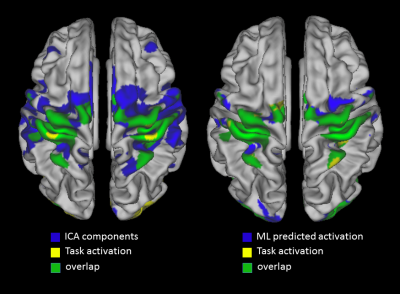
Figure 1 shows
the overlap (green) between the actual map (yellow) and the predicted map (blue),
for both the ICA (left) and ML (right) methods. In especially the motor cortex,
the predicated activation map demonstrates patterns more similar to the actual
motor activation derived from task fMRI.
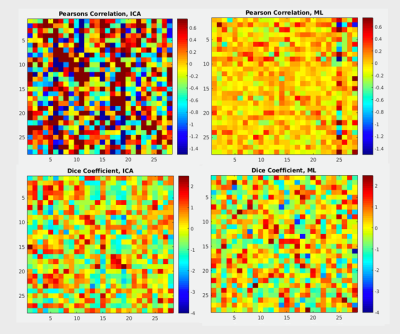
Figure 2: Pearson’s correlation coefficient (top row) and
Dice coefficient (bottom row) matrices comparing all subjects’ predicted and
actual task activation maps.
In both cases, a more diagonal pattern (higher correlation to self than to
other) is desired. Using the traditional ICA maps (left) as predictors does
give more high-value Pearson’s correlations, however these are not confined to
the diagonal elements as well as using the presented novel machine learning
prediction method (right). Dice coefficient values are similar in both
methods, but the matrix constructed using the novel model for predictions shows
more diagonality.