0865
Superselective 4D MR Angiography with Pseudo-Continuous Arterial Spin Labelling Combined with CENTRA-Keyhole (SS-4D-PACK) Used to Visualize Brain Arteriovenous Malformations1Clinical Radiology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan, 2Philips Japan, Tokyo, Japan, 3Philips Research, Hamburg, Germany, 4Division of Radiology, Department of Medical Technology, Kyushu University Hospital, Fukuoka, Japan
Synopsis
In the present study, we demonstrated the utility of superselective 4D-MR angiography with pCASL combined with CENTRA-keyhole (SS-4D-PACK) for the visualization of brain AVMs. This method enables a time-resolved and vessel-selective angiography within 5 minutes without a use of contrast agents. It was showed that almost perfect vessel selectivity was achieved with SS-4D-PACK. Although CNRs were slightly reduced in SS-4D-PACK due to a labeling loss during superselective label focusing, this was acceptable since visualization was well preserved. SS-4D-PACK can be a non-invasive clinical tool for assessing brain AVMs.
Introduction
Brain AVM is a vascular disorder that can cause serious intracranial hemorrhage, with an annual rate of rupture of 2%–3%1. DSA is the current reference standard for the diagnosis of brain AVM, however, it carries the risk of neurological complications and adverse reactions of contrast agents. Recently we proposed a new approach named 4D-pCASL-based angiography using CENTRA-keyhole (4D-PACK), which enables multi-phase acquisition within a short scan time by using keyhole and view sharing, while keeping a high flow signal in later phases by using pCASL2,3. Superselective-pCASL was introduced as a pCASL based vessel-selective labeling method4,5. On top of pCASL, superselective-pCASL uses additional gradients perpendicular to the flow direction and modifies the labeling efficiency in two directions. By dynamically changing the direction of the additional gradients in combination with changes to the phases of the RF pulses to neutralize the phase effects of these gradients at the targeted location, circular or elliptical labeling spots can be created.The size of the labeling focus can be adjusted by changing the zeroth moment of the transverse gradients; hence this method is able to label individual vessels. We combined superselective-pCASL with 4D-PACK for superselective 4D-MRA (SS-4D-PACK). In this study, we evaluated the utility of SS-4D-PACK for the visualization of brain AVMs by comparing with DSA and non-selective 4D-PACK.
Methods
[Patients] Five patients with brain AVM (age 41.5 ± 20.9 years; 4 males, 1 female) were examined. All patients underwent MRA and DSA within one month.
[MRI] MR imaging were performed on a 3T MR scanner (Ingenia 3.0T, Philips, Best, The Netherlands) with a 15-channel head coil.
SS-4D-PACK: Individual labeling of the right and left internal carotid artery (ICA), and the basilar artery was performed using the superselective-pCASL method. The labeling focus was placed in the upper cervical segment of ICA or lower basilar artery (Fig. 1). The gradient moment was set to be 0.75 mT/m msec to create a labeling spot with a diameter of approximately 2cm. Images were obtained by changing the label duration: 100ms, 200ms, 500ms, 800ms, 1200ms, 1600ms, 2000ms. Scans were further accelerated with view sharing. The other parameters were: sequence, 3D T1-TFE; TR/TE, 5.0/1.81ms; FA, 11°; ETL, 60; slab thickness, 80mm; voxel size, 1.0*1.4*1.6mm; SENSE, 3.0; keyhole 70%, acquisition time: approx. 5min.
4D-PACK: Images were obtained with label durations of 100ms, 200ms, 400ms, 600ms, 800ms, 1200ms, 1600ms, and 2200ms. Other parameters were the same as those for SS-4D-PACK.
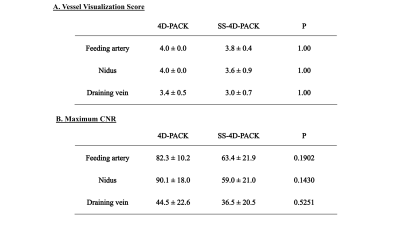
[Image Analysis] 1) The accuracy of vessel selectivity was assessed with a 4-point scale6 (1, poor; 4, excellent), 2) The visualization of the feeding artery, nidus, and draining vein was assessed using the DSA image as the reference standard with a 4-point scale; 3) The CNRs were measured using the following equation; CNR = (Vesselmax – WMave) / WMSD.
[Statistical Analysis] The visualization scores and maximum CNRs were measured between 4D-PACK and SS-4D-PACK using paired t-test.
Results
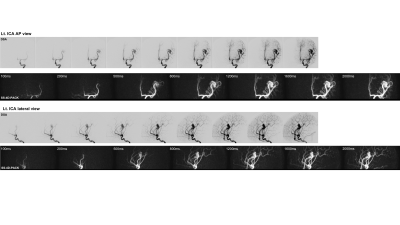
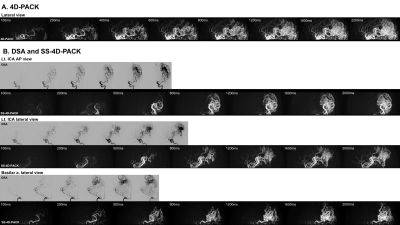
The vessel selectivity with SS-4D-PACK was perfect (grade 4) in 14 out of 15 territories (93.3%). A contralateral ICA territory was slightly visualized (grade 3) in one patient. Otherwise, no untagged vessels were observed. The ECA branches were not visualized in all patients. Figure 2 shows temporal changes in CNR for 4D-PACK and SS-4D-PACK. Figure 3 shows the visualization scores (A) and maximum CNRs (B). Maximum CNRs of all components tended to be lower in SS-4D-PACK than in 4D-PACK, but no significant differences were found. The visualization scores were not different between the two methods. Figure 4 shows an AVM fed by the left MCA branches and drained by the superficial middle cerebral veins. The selective labeling of the left ICA was successful, and no untagged vessels were observed. SS-4D-PACK sequentially depicted feeding arteries, nidus, and then draining veins, and its visualization was comparable to that on DSA. Figure 5 shows a large-sized AVM fed by multiple arteries from both the left MCA and left PCA. Conventional 4D-PACK (A) well visualized the whole lesion, but it was difficult to identify and separate multiple vessels. The SS-4D-PACK (B) enabled the separate labeling and imaging of the left ICA and basilar artery like DSA.Discussion
SS-4D-PACK enabled a time-resolved and vessel-selective angiography within 5 minutes without contrast agents. Almost perfect vessel selectivity was achieved with this method. Although CNRs were slightly reduced in SS-4D-PACK presumably due to labeling loss during superselective label focusing, this was acceptable since visualization was well preserved.Conclusion
SS-4D-PACK allowed for vessel-selective 4D-MRA and was useful in visualizing brain AVMs. SS-4D-PACK can be a non-invasive clinical tool for brain AVMs.Acknowledgements
No acknowledgement found.References
1. Hernesniemi JA, Dashti R, Juvela S, et al. Natural history of brain arteriovenous malformations: A long-term follow-up study of risk of hemorrhage in 238 patients. Neurosurgery 2008;63:823-829.
2. Obara M, Togao O, Okuaki T et al. Non-contrast enhanced 4D intracranial MR angiography based on pseudo-continuous arterial spin labelling (PCASL) with the keyhole technique. Proceedings of the 24th Annual Meeting of ISMRM, Singapore, 2016 #4376
3. Togao O, Hiwatashi A, Obara M, et al. 4D MR angiography with pseudo-continuous arterial spin labelling combined with CENTRA-keyhole (4D-PACK): visualization of distal cerebral arteries in Moyamoya disease. Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, 2017 #0145
4. Helle M, Norris DG, Rüfer S, et al. Superselective Pseudocontinuous Arterial Spin Labeling. Magn Reson Med 2010;64: 777–786.
5. Helle M, Rüfer S, van Osch MJ, et al. Superselective arterial spin labeling applied for flow territory mapping in various cerebrovascular diseases. J Magn Reson Imaging. 2013;38:496-503.
6. Fujima N, Osanai T, Shimizu Y et al. Utility of noncontrast-enhanced time-resolved four-dimensional MR angiography with a vessel-selective technique for intracranial arteriovenous malformations. J Magn Reson Imaging. 2016;44:834-45.
Figures
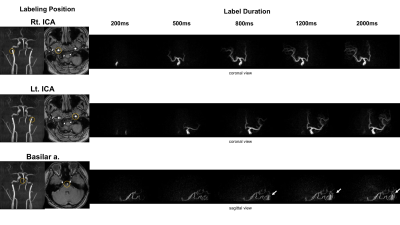
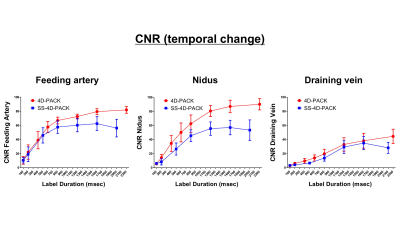
Figure 2. Temporal changes in CNR of feeding artery (left), nidus (middle), and draining vein (right) for 4D-PACK and SS-4D-PACK. CNRs of all components tended to be slightly lower in SS-4D-PACK than in 4D-PACK.