0833
Dynamic B0 Shimming for Multi-Slice Metabolite Mapping at Ultra-High Field in the Human Brain: Very High Order Spherical Harmonics vs. Multi-Coil1Max Planck Institute for Biological Cybernetics, Tuebingen, Germany, 2IMPRS for Cognitive and Systems Neuroscience, Eberhard-Karls University of Tuebingen, Tuebingen, Germany, 3Department of Biomedical Magnetic Resonance, Eberhard-Karls University of Tuebingen, Tuebingen, Germany, 4Department of Physics, Ernst-Moritz-Arndt University Greifswald, Greifswald, Germany
Synopsis
In this work, we evaluated the performance of two B0 shimming approaches (i.e. a very high order spherical harmonic shim system versus a multi-coil shim setup), for dynamic shimming in the context of high resolution and multi-slice metabolite mapping of the human brain at 9.4T.
Introduction
The many advantages of ultra-high field strengths for spectroscopy applications cannot be realized without a spatially homogenous B0 field. While commercial MR scanners are usually equipped with up to 2nd or 3rd degree spherical harmonic shim coils, recent studies1-4 have shown that for higher field strengths these shim coils are not sufficient. Two main hardware setups have been shown to be viable options for achieving better B0 shim quality at higher field strengths: shim systems with higher degrees of spherical harmonics shim coils2-4, or multi-coil setups1.
The aim of this work was to compare these two shim approaches in the context of 1H MRSI at 9.4T. For the first time, multi-slice and high resolution metabolite maps of the human brain recorded using slice-wise dynamically updated very high degree spherical harmonics and multi-coil B0 shimming are presented and compared.
Methods
All measurements were performed on a Siemens 9.4T whole-body human scanner with an in-house developed 18Tx/32Rx RF coil5. A very high order insert shim (VHOS) from Resonance Research Inc. with complete 4th and partial 5th- and 6th-degree spherical harmonic shim coils was used for very high-degree B0 shimming. A multi-coil6 (MC) shim array with 16 identical circular coils arranged in two rows was used for the localized multi-coil B0 shimming. The multi-coil setup was used as an additional layer of shimming after the ROI was primarily shimmed with the 1st and 2nd-order/degree spherical harmonic shim coils from the vendor. For a fair comparison, only the 3rd and 4th-degree shim coils from the insert shim were used, so that the total number of channels of VHOS system was also 16 channels. Therefore, both methods used up to 2nd-order/degree shimming and 16 additional coils.
For the VHOS system, 4th degree static shimming (S4) was compared to 2nd (D2) and 4th (D4) degree dynamic slice-wise shimming. For the MC system, slice-wise shimming using the multi-coil setup (DMC) was compared to 2nd degree slice-wise shimming (D2).
Additionally, each shim setup was used for multi-slice high resolution metabolite mapping from three slices on the brains of healthy volunteers. A non-lipid-suppressed 1H FID MRSI sequence7 with the following parameters was used for metabolite mapping: TR=300ms, TE=1.5ms, slice thickness=10mm, 100% distance factor, matrix size=64x64, FOV=200x200mm, spectral bandwidth 8kHz.
Results/Discussion
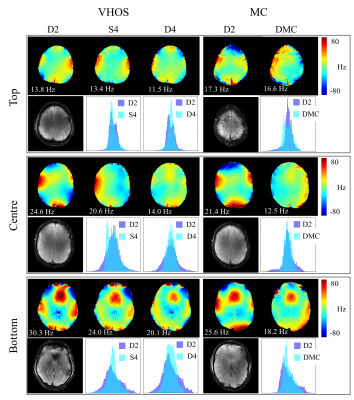
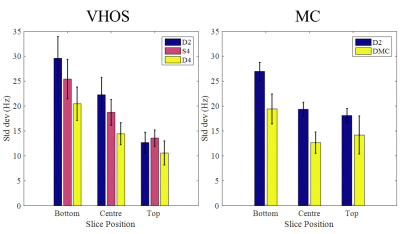
Figure 1 shows the results of in-vivo B0 shimming comparisons using the two different shim systems. The overall performances (averaged over 3 volunteers) are summarized in Figure 2.
Comparison of static and dynamic shimming with the VHOS system shows that on average 4th order static shimming outperforms 2nd order dynamic shimming for the centre and bottom slices. However, the improvement is traded-off for the homogeneity of the top slice.
The improvement from D2 to D4 is comparable to the improvement seen from D2 to DMC. However, the improvement is always less for the top slices compared to the centre and bottom slices.
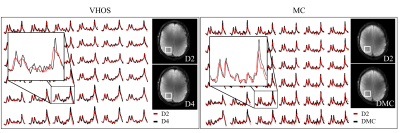
Figure 3 shows representative spectra for the different B0 shimming settings. Two different comparisons are represented: spectra acquired with D2 vs. D4 and once again D2 vs DMC. As expected, the more homogeneous the B0 field, the narrower the linewidths of the spectra.
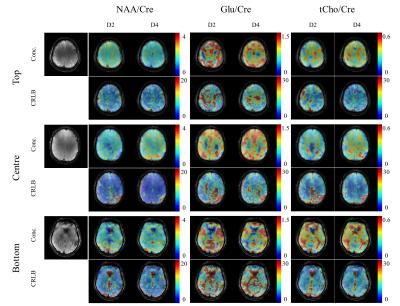
The resulting metabolite maps along with their respective Cramer-Rao lower bound (CRLB) maps for the VHOS comparisons are shown in Figure 4. For NAA on the bottom slice the signal dropout due to the residual B0 inhomogeneity from the nasal cavity is smaller when using D4 than when using D2. Also, in the CRLBs of NAA, more voxels could be reliably fit for the centre and bottom slices when D4 shimming was used. The effect of good shimming is more obvious for the Glutamate and total Choline metabolite maps.
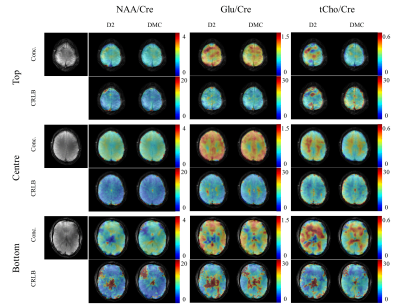
The resulting metabolite and CRLB maps for the MC comparisons are shown in Figure 5. Here, we can see very good improvement of the DMC over D2 (particularly for the bottom slice). Again, the metabolite maps resemble the underlying anatomical structure better for the DMC shimming. Furthermore, the improved fitting is also clearly reflected in the CRLB maps for the bottom slice and the total Choline in the top slice.
Conclusion
Both the VHOS and the MC setup showed advantages over low degree (up to 2nd) spherical harmonic dynamic shimming for multi-slice and high resolution metabolite mapping and resulted in more anatomically accurate maps. These results emphasize the importance of slice-wise B0 shimming for metabolite mapping at ultra-high fields and suggest that either of the two B0 shimming approaches can be used to achieve similar results.Acknowledgements
No acknowledgement found.References
[1] Juchem et al, MRM 2015
[2] Boer et al, MRM 2014
[3] Pan et al, MRM 2012
[4] Chang et al, MRM 2017
[5] Avdievitch et al, NMR in Biomed 2017
[6] Aghaeifar et al, ISMRM 2016
[7] Nassirpour et al, NeuroImage 2016
Figures