0798
Multicenter and multiscanner reproducibility of Magnetic Resonance Fingerprinting relaxometry in the brain1Siemens Healthcare GmbH, Erlangen, Germany, 2Max Schaldach-Stiftungsprofessur für Biomedizinische Technik, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 3Siemens Medical Solutions USA Inc., Malvern, PA, United States, 4Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 5Radiology, Case Western Reserve University, Cleveland, OH, United States
Synopsis
Magnetic Resonance Fingerprinting (MRF) is a technique for generating tissue property maps by matching pseudo randomly varying MR signal time courses in each voxel with a precalculated dictionary of possible evolutions. In this study, the repeatability of MRF results on the same scanner as well as the reproducibility on different scanners is evaluated for the human brain.
Purpose
Magnetic Resonance Fingerprinting (MRF)1 is a new technique that promises multi-parametric, quantitative MRI. A series of RF pulses generates a complex signal response in non-steady-state which is compared to a physically modelled signal to determine the properties of interest in the tissue from which the signal originates.
This study was conducted to evaluate the repeatability of MRF results on the same scanner as well as the reproducibility on different scanners in brains of healthy volunteers.
Methods
The study employed a prototype implementation of FISP-MRF2.To mitigate the influence arising from inhomogeneities of the transmit RF, a B1+ correction was used. Here a B1+ prescan3 is acquired, and the relative B1+ value is used to select a subdictionary for the matching process, calculated with the corresponding relative B1+. The spiral trajectory for sampling k-space was rotated by 82.5° for succeeding TRs to mitigate undersampling artifacts4,5.
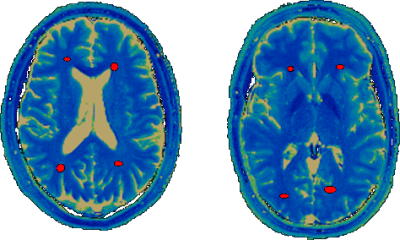
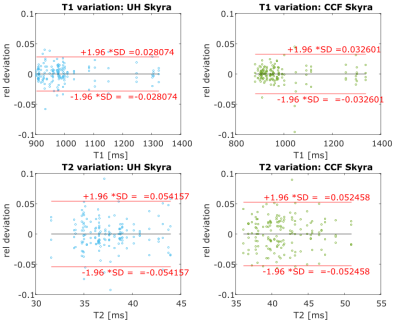
During one week, four healthy volunteers’ brains (age 23 to 47 years, 3 male, 1 female) were scanned on four different 3-Tesla scanners in Erlangen, Germany (2x MAGNETOM Skyra, MAGNETOM Prisma, MAGNETOM Prismafit, Siemens Healthcare, Erlangen, Germany). Two months later two of the same volunteers were also scanned on two MAGNETOM Skyra systems at two different and unrelated facilities in Cleveland, OH, USA. The protocol was set up to cover 7 slices with an in-plane resolution of 1.17 mm and 5mm thickness in a total scan time of 5 minutes. All volunteers were scanned four times on each scanner during one session, each repetition being preceded by an Auto-Align scout and subsequent automated repositioning of the slices. To minimize the influence of varying slice angulation and position, elliptical ROIs consisting of 20 to 40 pixels were drawn in two white-matter regions bilaterally in each slice (Examples are shown in Figure 1). A mean T1 and T2 value was calculated for each volunteer, slice and scanner. The relative deviations of these values from the means over all scanners are displayed as Bland-Altman plots showing the inter-scanner variation. The variations over repetitions per scanner were analogously calculated and displayed as Bland-Altman plots.
Results
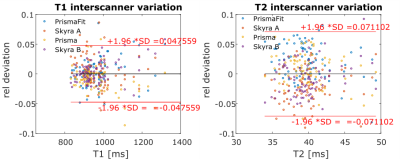
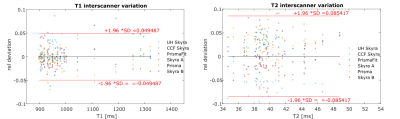
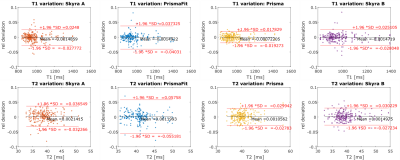
The inter-scanner variation of T1 and T2 in Erlangen, Germany, based on the data of all four volunteers is displayed in figure 2. 95% of T1 values deviate less than 4.8% from the mean and 95% of T2 values deviate less than 7.1% from the mean. The inter-scanner variation of T1 and T2 of all scanners in Germany and USA, based on the data of two volunteers is displayed in figure 3. 95% of T1 values deviate less than 5% from the mean, and 95% of T2 values deviate less than 8.5% from the mean. Intrascanner variation is displayed in figure 4 for scanners located in Erlangen, Germany and in figure 5 for scanners in Cleveland, OH, USA.Discussion
Previous studies7 have shown MRF reproducibility on phantoms. In this work, we show that also T1 and T2 values in healthy volunteer brains measured with MRF are highly reproducible over different scanners, sites, and time. It should be noted that deviations not solely arise from the method itself but are also affected by positioning differences of the ROIs, either caused by head motion or changes in head position between different scanner sessions. For mitigation, each scan was positioned based on the Auto-Align prescan with land mark detection. Another potential influence for the higher inter-scanner variation is physiological changes in the brain tissues that even lead to a varying morphology6. Especially long travels and slightly different times of day when scans were conducted can be expected to affect results.Conclusion
In this study we provide numbers for reproducibility and repeatability of Magnetic Resonance Fingerprinting relaxation parameter maps in a controlled experiment on healthy volunteers’ brains over multiple scanners.Acknowledgements
No acknowledgement found.References
1 Ma D. et al, Magnetic resonance fingerprinting. Nature 2013;
2 Jiang Y. et al, MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. MRM. 2015;
3 Chung S. et al., Rapid B1+ mapping using a preconditioning RF pulse with TurboFLASH readout, MRM 2010;
4 Pfeuffer, J. et al, Mitigation of Spiral Undersampling Artifacts in Magnetic Resonance Fingerprinting (MRF) by Adapted Interleaf Reordering. ISMRM 2017;
5 Körzdörfer, G. et al, Spatial biases in Magnetic Resonance Fingerprinting parameter maps arising from undersampling patterns. ISMRM 2017;
6 Trefler A et al, Impact of time-of-day on brain morphometric measures derived from T1-weighted magnetic resonance imaging, Neuroimage 2016
7 Jiang Y et al, Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom, MRM 2016
Figures