0772
Optimized respiratory-resolved motion-compensated 3D Cartesian coronary MRA1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom
Synopsis
Conventionally, free-breathing whole-heart 3D coronary MR angiography (CMRA) uses navigator-gated acquisitions to reduce respiratory motion, by acquiring data only at a specific respiratory phase, which leads to prolonged scan times. Respiratory-resolved reconstruction approaches have been proposed to achieve 100% scan efficiency using mainly non-Cartesian acquisitions and exploiting sparsity in the respiratory dimension. Here, a robust framework for Cartesian imaging is proposed, which provides high-quality respiratory-resolved images by incorporating motion information from image navigators (iNAV) to increase the sparsity in the respiratory dimension. Furthermore, iNAV motion information is used to compensate for 2D translational motion within each respiratory phase.
Introduction
A major challenge in free-breathing whole-heart coronary MR angiography (CMRA) is image degradation due to respiratory motion. Diaphragmatic navigator-gated acquisitions are commonly used to minimize respiratory motion. However, this approach leads to prolonged acquisition times, since only data within a small end-expiration gating window is accepted. Moreover, it ignores the significant pathophysiological interactions between the cardiovascular and respiratory systems.1 Recently, XD-GRASP has been proposed to achieve 100% scan efficiency (no data rejection) and provide respiratory-resolved 3D radial CMRA images.2,3 Most respiratory-resolved approaches2-4 sort the acquired data into multiple respiratory phases (or bins) and reconstruct the bin images by exploiting Total Variation (TV) sparsity along the respiratory dimension. However, for Cartesian liver imaging XD-GRASP has been shown to suffer from reduced image quality, especially for bins with large respiratory displacements.5 Here, a robust framework for Cartesian imaging is proposed to provide high quality respiratory-resolved CMRA images. The proposed Optimized Respiratory-resolved Cartesian Coronary MR Angiography (XD-ORCCA) reconstruction incorporates motion information to increase the sparsity in the respiratory dimension (similarly to methods proposed for cardiac motion6). Furthermore, 2D beat-to-beat translational motion information extracted from image navigators7 (iNAVs) is used to minimize intra-bin motion. The proposed XD-ORCCA was tested on seven healthy subjects and compared against XD-GRASP.Methods
3D CMRA data is acquired using a prototype golden-step spiral-like Cartesian (CASPR) trajectory,8 which samples the ky-kz plane with spiral interleaves on a Cartesian grid. Consecutive spirals are separated by the golden-angle. A low-resolution 2D iNAV is acquired in every heartbeat, before each spiral interleaf of the whole-heart 3D CMRA acquisition. The 2D iNAVS are registered to a common respiratory position (end-expiration) to estimate beat-to-beat 2D translational (right-left and superior-inferior: SI) motion. The estimated SI motion is used to group the 3D CMRA data into five equally populated respiratory bins. In XD-ORCCA, 2D translational motion correction within each bin is performed in k-space before the reconstruction, thereby improving image quality of each bin. During the reconstruction, intra-bin motion corrected images are aligned to one respiratory position to increase sparsity in the respiratory domain.
Each undersampled bin is reconstructed using: 1) XD-GRASP and 2) XD-ORCCA. The respiratory-resolved images were obtained by solving the following optimization problems: 1) $$$\hat{\rm{\bf{x}}}=\rm{arg} \min\limits_{x}\left\{ \frac{1}{2}\left\|\mathbf{E}\,\mathbf{x}\,-\mathbf{k}\right \|_2^2+\alpha\,\Psi_t(\mathbf{ x})\right\}$$$ and 2) $$$\hat{\rm{\mathbf{x}}}=\rm arg\min\limits_{x}\left\{\frac{1}{2}\left\|\bf E\,\mathbf{x}\,-\mathbf{b}\right\|_2^2+\alpha\,\Psi_t(\mathcal{R}\mathbf{x})+\beta\,\Psi_s(\mathbf{x})\right\}$$$, where $$$\bf{x}$$$ is the respiratory-resolved image series, $$$\bf{k}$$$ is the binned k-space data, $$$\bf{b}$$$ is the 2D translational corrected binned k-space data, $$$\Psi_{\rm s}$$$ is the 3D spatial TV function, $$$\alpha$$$ and $$$\beta$$$ are regularization parameters, $$$\mathcal{R}\mathbf{x}=T_{i}\mathbf{x}_i$$$ is the motion-corrected domain, where $$$T_i$$$ is the translation transform that maps the bin image $$$\mathbf{x}_i$$$ to the reference image $$$\mathbf{x}_1$$$ (end-expiration), and $$$\Psi_{\rm t}=\mathbf{x}_1-T_{i}\mathbf{x}_i$$$ is the 1D temporal TV function. The operator $$$\mathbf{E = AFS}$$$ incorporates the sampling matrix $$$\bf{A}$$$ for each bin, Fourier transform $$$\bf{F}$$$ and coils sensitivities $$$\bf{S}$$$. Additionally, a 3) XD-GRASP with intra-bin translational motion correction (TC) representative reconstruction was obtained, which consisted in solving 3) $$$\hat{\rm{\bf{x}}}=\rm{arg}\min\limits_{x}\left\{ \frac{1}{2}\left\|\mathbf{E} \,\mathbf{x}\,-\mathbf{b}\right \|_2^2+\alpha\,\Psi_t(\mathbf{ x})\right\}$$$. These problems were solved with the nonlinear conjugate gradient method.
In-vivo free-breathing experiments were performed on seven healthy subjects on a 1.5T scanner (Siemens Magnetom Aera) with 18-channel body and 32-channel spine coils. 3D CMRA bSSFP acquisitions were performed using the following parameters: coronal orientation, FOV=320x320x80-104mm3, resolution=1x1x2mm3, TR/TE=3.6/1.56ms, flip angle=90°, T2 preparation (40ms), SPIR-like fat saturation, subject specific mid-diastolic trigger delay, acquisition window ~100ms, 1 spiral interleaf per R-R interval, with acquisition time ~9-12min. For the 2D iNAV acquisition, 14 bSSFP startup echoes were used (same geometry).
Results
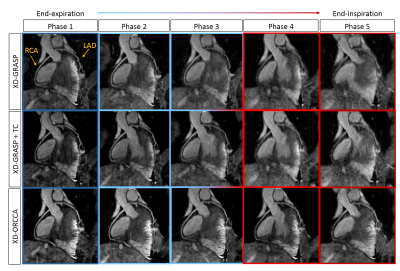
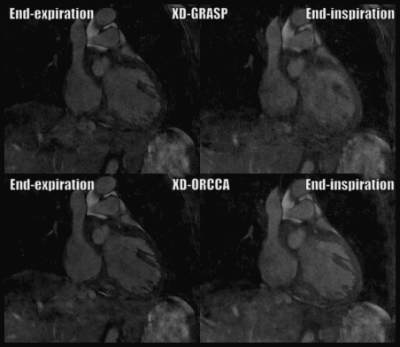
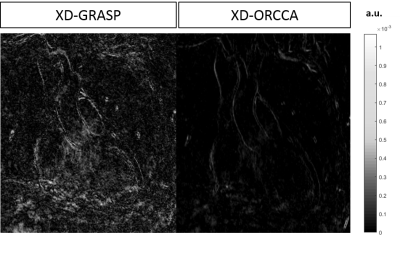
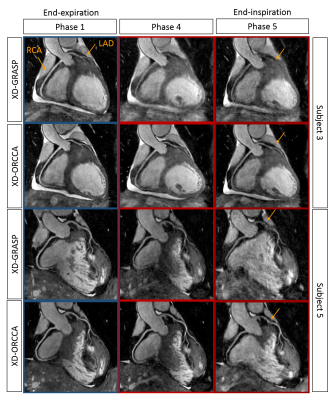
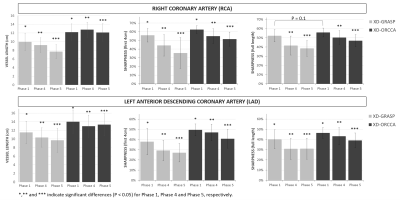
Reformatted and coronal respiratory-resolved images obtained with the proposed XD-ORCCA and XD-GRASP are shown in Figs.1-2 for a representative healthy subject. Motion blurring is observed in the XD-GRASP images, particularly for respiratory phases (3 to 5) in which the displacement of the heart is the largest. Fig.1 also shows that intra-bin TC slightly reduces motion in XD-GRASP, however blurring is still observed in phases 3 to 5. The proposed XD-ORCCA produces high-quality respiratory-resolved images, by increasing temporal sparsity (Fig. 3). Fig. 4 shows three reformatted respiratory phases obtained with XD-GRASP and XD-ORCCA for two subjects. Significant differences in average vessel length and sharpness were observed between XD-GRASP and XD-ORCCA for both coronaries for end-inspiratory phases (Fig. 5).Conclusions
A robust and highly efficient respiratory-resolved motion-compensated framework for 3D Cartesian CMRA has been proposed. Translational motion is estimated from 2D iNAV images and incorporated into the XD-ORCCA reconstruction to increase the sparsity in the respiratory dimension. Additionally, this motion information is used to compensate for residual intra-bin motion. The proposed XD-ORCCA provides high-quality respiratory-resolved images for all respiratory phases.Acknowledgements
This work was supported by the following grants: EPSRC EP/N009258/1, EP/P032311/1 and EP/P001009/1, EP/P007619/1 and FONDECYT N° 1161051.References
1. Remetz M, Cleman M, Cabin H. Pulmonary and pleural complication of cardiac disease. Clinics in Chest Medicine 1989; 10:545-592.
2. Feng L, Axel L, Chandarana H, et al. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimension using compressed sensing. MRM 2016; 75:775-788.
3. Piccini D, Feng L, Bonanno G, et al. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. MRM 2017; 77:1473-1484.
4. Cruz G, Atkinson D, Buerger C, et al. Accelerated motion corrected three-dimensional abdominal MRI using total variation regularized SENSE reconstruction. MRM 2016, 75:1484-1498.
5. Feng L, Chandarana H, Zhao T, et al. Golden-angle sparse liver imaging: radial or Cartesian sampling?. ISMRM 2017; #1285.
6. Asif M, Hamilton L, Brummer, Romberg J. Motion-adaptive spatio-temporal regularization for accelerated dynamic MRI. MRM 2012; 70:800-812.
7. Henningsson M, Koken P, Stehning S, et al. Whole-heart coronary MR angiography with 2D self-navigated image reconstruction. MRM 2012; 67:437-445.
8. Prieto C, Doneva M, Usman M, et al. Highly efficient respiratory motion compensated free-breathing coronary MRA using golden-step Cartesian acquisition. JMRI 2015; 41:738-746.
Figures