0767
Improved phase unwrapping algorithm for automatic cine DENSE strain analysis using phase predictions and region growingDaniel Auger1, Xiaoying Cai1, Changyu Sun1, and Frederick Epstein1,2
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Radiology, University of Virginia, Charlottesville, VA, United States
Synopsis
Displacement encoding with stimulated echoes (DENSE) measures myocardial displacements using the signal phase. Phase wrapping generally occurs during systolic phases, thus spatiotemporal phase unwrapping algorithms are required to compute motion trajectories and strain. Current DENSE analysis methods are aided by user-defined myocardial contours. A fully automatic DENSE analysis method is proposed where phase predictions using multiple pathways and region growing are used to simultaneously unwrap and segment the myocardium. Compared to a prior automatic method, this method selects fewer extramyocardial pixels, reducing the computation time, and has a greater phase unwrapping success rate.
Introduction
Cine DENSE measures myocardial displacements by encoding tissue displacement into the signal phase. Displacement encoding frequencies (ke) are selected to balance signal-to-noise ratio, displacement sensitivity, and artifact suppression [1], resulting in phase wrapping during systole. Spatiotemporal phase unwrapping is required to compute Lagrangian motion trajectories and strain [1, 2]. Phase unwrapping may be aided by delineating the myocardium using manually-defined contours [3]. A fully automatic phase unwrapping method would eliminate the need for user intervention, and a method was previously developed, however it identified many extramyocardial pixels as input into the displacement trajectory and strain calculations [2], increasing the computational burden. A further limitation of the previous algorithm was an inability to unwrap DENSE datasets with multiple cycles of wrap. We propose a phase unwrapping algorithm based on phase predictions using multiple pathways and region growing [4]. This approach may identify fewer extramyocardial pixels and successfully unwrap datasets with multiple cycles of phase wrap.Methods
The proposed method utilizes multiple phase prediction pathways and consistency checking for region growing. Initially, a small myocardial region is automatically identified by applying principle component analysis to DENSE images. The initial region is selected at an early cardiac phase where phase wrapping has not occurred. A perimeter of potentially phase-wrapped pixels around the initial region is identified, and multiple spatiotemporal linear predictions from previously unwrapped pixels are used to predict the phases of the perimeter pixels (Fig 1(A)). Once an attempt is made to unwrap a pixel, reliability thresholds are applied to assess the attempt. The thresholds are based on the overall predicted phase (TPrediction) and the variation of the multiple predictions (TDeviation). If the attempt is deemed reliable, the pixel is included in the unwrapped region. The process continues, allowing the region to grow and including regions with reliable predictions and rejecting regions with unreliable predictions, i.e., noise. For 2D displacement encoding, the algorithm is simultaneously applied to both encoding directions so that the region grows identically for both dimensions. The prior automatic unwrapping algorithm [2] and our proposed algorithm were applied to 5 volunteer datasets acquired using 2D short-axis cine DENSE with localized signal generation and with ke = 0.1 cyc/mm, in-plane pixel size of 2.3 × 2.3 mm2, and 24-32 cardiac phases. To generate gold standard data for comparisons, the myocardium was manually contoured, phase unwrapping was applied to the segmented region, and the unwrapping results were manually inspected to ensure the absence of unwrapping errors. Optimal thresholds were determined by using the new algorithm with a range of thresholds and finding those that minimize the root mean square (RMS) error between the gold-standard data and the masked results from the proposed algorithm. To test the algorithm, additional datasets (n=3) were acquired with displacement encoding frequencies of 0.14, 0.18 and 0.22 cyc/mm, leading to increased degrees of phase wrapping. Displacement trajectories and principle shortening strain (PSS) were calculated using the proposed method and the gold standard method [3, 4].Results
Fig 1(B) illustrates RMS error plots for all ke values to identify the best reliability thresholds. The unsmooth RMS error function is due to the non-linearity of the phase-unwrapping operation. Optimal values of π /3 for TPrediction and 7π /12 for TDeviation were identified. Fig. 1(C, D) illustrate poor phase unwrapping where either TPrediction and TDeviation were too low and regions of myocardium were improperly excluded during region growing, or TPrediction and TDeviation were too high and regions of surrounding tissue and blood were improperly included in the growth region and caused unwrapping errors. Fig. 1(E) illustrates successful region definition and myocardial phase unwrapping using the optimal thresholds. Fig. 2 shows that the proposed algorithm excludes more extramyocardial pixels than the prior automatic method. Fig. 3 illustrates that the proposed algorithm successfully unwrapped datasets with multiple cycles of phase wrapping, in contrast to the prior algorithm. For different ke values, Fig. 4 illustrates the decrease in incorrectly unwrapped frames using the proposed algorithm compared to the prior algorithm. Fig 5(A) shows an end-systolic PSS map automatically computed using the proposed method. Fig 5 (B-E) show the comparison of PSS computed automatically and using manually-delineated myocardial contours, and Fig 5(D, E) show the Bland-Altman and correlation plots comparing the two methods indicating a small bias (+0.007) and excellent correlation (R2=0.82).Conclusion
Fully automatic phase unwrapping using multiple phase prediction pathways and region growing provides better delineation of myocardial tissue and more reliable phase unwrapping than prior automatic methods. The proposed method may facilitate fully automatic in-line strain mapping for DENSE in the future.Acknowledgements
Research support from Siemens HealthineersReferences
References: 1. Spottiswoode. IEEE TMI 2007; 26:15-30 2. Gilliam. IEEE TMI 2012; 31:1669-81. 3. Spottiswoode. MIA 2009; 13:105–115 4.Xu. IEEE TGRS 1999; 37:124-34Figures
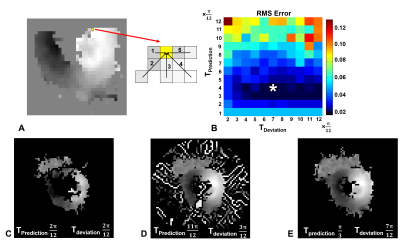
Figure 1: (A) Use of multiple prediction pathways to automatically
phase unwrap a neighboring pixel. (B) Phase-unwrapped RMS error plots, based on
manually-contoured phase-unwrapped images as the gold standard, for all ke
values (0.1, 0.14, 0.18, 0.22) and threshold values. The optimal thresholds correspond
to the pixel indicated by *. (C) Unwrapping result using low thresholds lead to
the improper exclusion of myocardial pixels. (D) Unwrapping results using high
threshold values lead to the improper inclusion of surrounding tissue and blood
and to subsequent phase unwrapping errors. (E)
The optimal thresholds lead to identification of all myocardial tissue
and correct phase unwrapping.
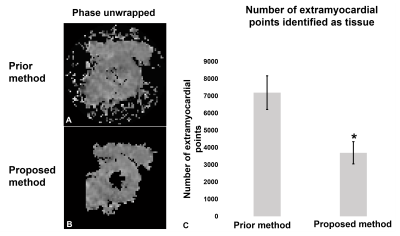
Figure 2: (A, B) Phase
unwrapping results show that the proposed method excludes more
extramyocardial pixels compared to the previous automatic method. (C) For all
volunteers, the proposed method significantly reduced the number of pixels used
for displacement and strain calculation thus reducing the overall computation
time. * P<0.01
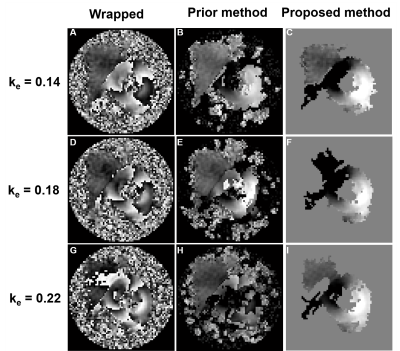
Figure 3: Examples of phase unwrapping from DENSE datasets
with increasing values of ke resulting in multiple cycles of
wrapping. The proposed method successfully unwrapped the myocardium for all ke
values whereas the prior phase unwrapping algorithm failed. Also, the proposed
method removed more of the surrounding tissue compared to the previous method.
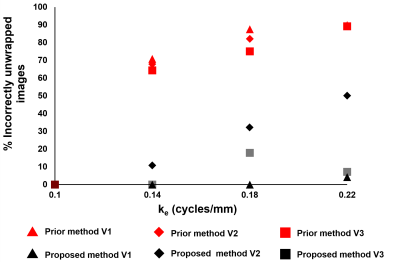
Figure 4: The percent of phase unwrapping errors for both
methods at varying ke values for 3 volunteers. The proposed method
shows a reduced number of incorrectly unwrapped images for all ke
values. Vi stands for volunteer “i".
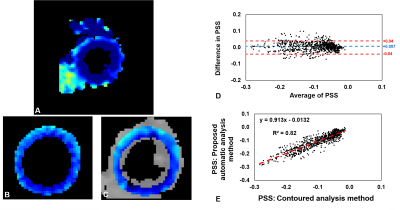
Figure 5: Agreement and correlation between PSS calculated
using manually-drawn LV contours and automatically. (A) End-systolic PSS map
computed automatically in part using the proposed phase unwrapping algorithm. (B)
PSS calculated using manually-drawn contours.
(C) PSS calculated automatically and subsequently masked with manually-drawn
contours. (D) Bland-Altman plot of segmental PSS for all volunteers using data
from the masked region. (E) Correlation plot of segmental PSS for all
volunteers from the masked region.