0677
MR Fingerprinting for High Resolution Metabolic Imaging of Hyperpolarized [1-13C]Pyruvate1Department of Radiology & Biomedical Imaging, UCSF, San Francisco, CA, United States, 2GE Healthcare, San Francisco, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States
Synopsis
The purpose of this study was to investigate a new application of MR fingerprinting (MRF) methods for hyperpolarized 13C MRI. We show that introducing randomization of pulse parameters into a 13C SSFP acquisition train facilitates efficient extraction of individual hyperpolarized 13C metabolite levels based on their transient signal responses (i.e. "fingerprints"), in simulations as well as phantom and in vivo experiments. Application of MRF methods in this multi-spectral SSFP framework enables exploitation of the long T2 relaxation times of 13C nuclei for increased sensitivity and spatial resolution in hyperpolarized MRI. Initial results show that MRF approaches have great potential for application to hyperpolarized 13C metabolic imaging.
Introduction
The massively increased sensitivity provided by hyperpolarization has been shown to support HP 13C imaging at spatial resolutions approaching one millimeter isotropic, using refocused SSFP acquisition1. Prior “metabolic imaging” studies (e.g. for imaging conversion of hyperpolarized pyruvate to lactate) have however been limited to much coarser spatial resolutions2 (~3-5mm). Refocused SSFP methods exploit the long T2's of 13C nuclei for improved sensitivity and spatial resolution1, but managing the spectral response of SSFP trains is difficult in the context of metabolic imaging, which involves systems with multiple resonances3. In this work we show initial results from the application of a randomized magnetic resonance fingerprinting4 (MRF) approach to efficiently extract inidividual hyperpolarized 13C metabolite levels based on the characteristic transient SSFP signal evolutions (i.e. “fingerprints”) of individual resonances.Methods
Inspired by the recent development of randomized MRF methods for rapid feature detection from transient signal evolutions in conventional 1H MRI4 (e.g. proton density, T1, B0, etc.), we recently found that introducing randomization into 13C SSFP acquisition trains similarly greatly improves the conditioning of multi-spectral reconstruction of hyperpolarized 13C metabolic imaging data.
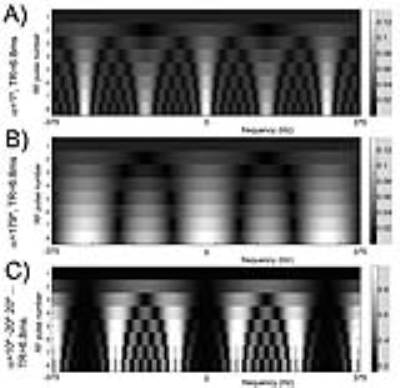
Bloch simulations- We simulated the transient signal responses of the principal metabolites in the hyperpolarized [1-13C]pyruvate system (i.e. pyruvate, alanine, pyruvate hydrate, and lactate) to arbitrary SSFP pulse trains. For example, conventional transient SSFP profiles are shown in Fig. 1.
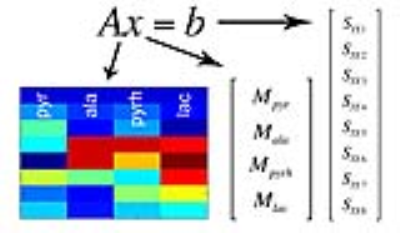
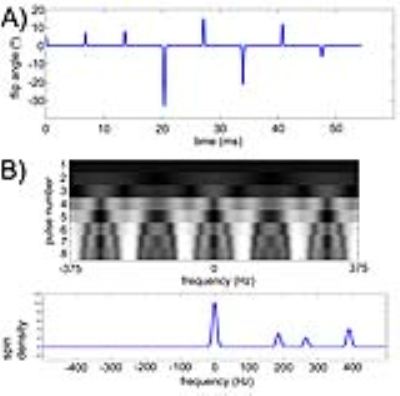
Design of MRF acquisition train- A key requirement for the proposed multi-spectral MRF is that the time-evolutions (“fingerprints”) are distinct between HP [1-13C]pyruvate and its metabolic products. In other words, the multi-spectral signal encoding matrix A, as defined in Fig. 2, must be well-conditioned in order to solve for the individual metabolites. An initial “proof of concept” implementation of a randomized approach for 13C MRF, derived from empirically varying the flip angle train, is illustrated in Fig. 3. A minimum TR of 6.4ms was enforced, as required for 1.5mm resolution along the readout dimension on our 3T scanner. The computed magnetization profile (Fig. 3A) is seen to be spectrally spread out by randomization of the flip angle train, as compared with the standard profiles shown in Fig. 2. As this occurs, increasingly unique signal responses for the individual metabolites are generated. The condition number of this particular A matrix is ~2 (unity= ideal noise performance).
MRI experiments- Spin density for all four metabolites (0.3ppm linewidth for each, as shown in Fig. 3B) was applied to the above magnetization profile in order to calculate predicted MRI signal evolutions. These computed responses were compared against results obtained by imaging a 13C phantom (a vial containing 6M [13C]urea) at transceiver offset frequencies corresponding to the four metabolites. Corresponding metabolite images were also reconstructed for each of these offsets. For this initial study, source components were recovered by direct matrix inversion. Finally, an initial in vivo imaging study in a rat was also conducted for proof of principle. The rat was injected with 2.2mL 80mM hyperpolarized [1-13C]pyruvate via tail vein, with 1.5mm x 1.5mm coronal projection MRF imaging initiated 20s after injection.
Results
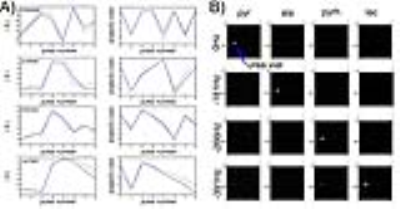
Phantom studies- As shown in Fig. 4, acquired signal curves from a [13C]urea phantom, acquired at varying offset frequencies to simulate the hyperpolarized [1-13C]pyruvate system, closely match the predicted signal evolutions (i.e. the dictionary entries). This correspondence indicates that our simulation accounts for relevant experimental factors. This framework supported successful multi-spectral reconstruction of phantom image data as shown in Fig. 5, with spectral selectivity >10:1.
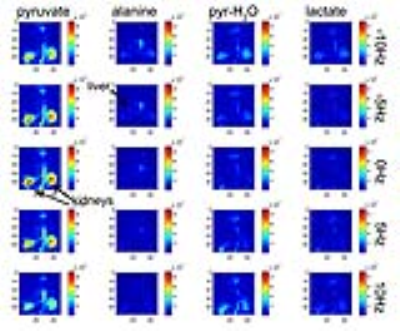
In vivo study- Reconstructed metabolite images from the in vivo rat experiment are shown in Fig. 5. Since the signal evolution and therefore the encoding matrix depends on local B0 offset, reconstructions are shown over a range of 20Hz (0.6ppm) B0 offset frequencies. Optimal reconstruction is a voxel-by-voxel combination of reconstructions executed over a range of B0 offset frequencies.
Discussion
The results of this study clearly show the potential for applying a randomized MRF approach for rapid multi-spectral hyperpolarized 13C imaging. Signal evolutions in vivo are, however, sensitive to local B0 offset-- this effect could be incorporated into the encoding matrix using B0 maps. Ultimately, exact reconstructions that account for additional parameters including B0, T1, and T2 will be determined by a dictionary-matching based approach.
The described multi-spectral MRF approach provides some flexibility over existing methods for multi-spectral MRI, such as chemical shift imaging (CSI) or multi-echo Dixon methods, which rely exclusively on magnetization phase evolution. The multi-spectral MRF approach utilizes both magnitude and phase of the hyperpolarized signal evolution, and thus is particularly adapted for well-conditioned feature detection in the transient state based on a limited set of measurements.
Acknowledgements
This work was supported by NIH K01DK099451 and P41EB013598.References
1. Reed GD, von Morze C, Verkman AS, Koelsch BL, Chaumeil MM, Lustig M, Ronen SM, Bok RA, Sands JM, Larson PE, Wang ZJ, Ardenkjaer-Larsen JH, Kurhanewicz J, Vigneron DB. Imaging Renal Urea Handling in Rats at Millimeter Resolution Using Hyperpolarized Magnetic Resonance Relaxometry. Tomography. 2:125-137 (2016).
2. Larson PE, Bok RA, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C]pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magnetic Resonance in Medicine. 63: 582-91 (2010).
3. von Morze C, Sukumar S, Reed GD, Larson PE, Bok RA, Kurhanewicz J, Vigneron DB. Frequency-specific SSFP for hyperpolarized 13C metabolic imaging at 14.1T. Magnetic Resonance Imaging. 31:163-70 (2013).
4. Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 495: 187-92 (2013).
Figures