0671
Comparison of cervical cord results from a quantitative 3D multi-parameter mapping (MPM) protocol of the whole brain with a dedicated cervical cord protocol1Queen Square MS Centre, University College London, London, United Kingdom, 2Spinal Cord Injury Center Balgrist, University of Zurich, Zurich, Switzerland, 3Neurophysics Department, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 4NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, QC, Canada, 5Functional Neuroimaging Unit, CRIUGM, Université de Montréal, Montreal, QC, Canada, 6Wellcome Trust Centre for Neuroimaging, UCL Institute of Neurology, University College London, London, United Kingdom, 7Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 8Brain MRI 3T Centre, C. Mondino National Neurological Institute, Pavia, Italy
Synopsis
We present a comparison of cervical cord metrics obtained using both a whole brain and dedicated cord implementation of a recently introduced quantitative multi-parametric MRI protocol which provides apparent proton density, R1, magnetisation transfer saturation (MTsat) and R2* maps sensitive to microstructural tissue changes in brain and spinal cord. Similar whole cervical cord (levels C1-C5) parameters were obtained using either protocol, and inter-subject variation was low, however in order to investigate tissue-specific cord parameters the dedicated cord protocol with higher in-plane resolution would be desirable.
Introduction
The spinal cord is commonly affected in neurological disorders such as multiple sclerosis, amyotrophic lateral sclerosis, cervical spondylotic myelopathy (CSM), and spinal cord injury(SCI)1, however performing quantitative tissue-specific cord MRI can prove challenging due to its small cross-sectional size and potential for cord motion. Multi-parameter mapping (MPM) is a quantitative imaging technique utilising multi-echo 3D spoiled gradient echoes (SPGR) (and B1 mapping for correction of T1 values) to quantify several parameters including apparent proton density (APD), R1 (=1/T1), R2* (=1/T2*), and Magnetisation Transfer saturation (MTsat)2-7. We evaluated results obtained in the cervical cord (C1-C5) using 1) a whole brain MPM protocol with cord coverage to at least level C5, and 2) a dedicated cervical cord MPM protocol (potentially allowing higher in-plane resolution and coverage for a shorter acquisition time), with a view to applying the method in patients with CSM or SCI, above the injury level.
Materials and Methods
MRI Acquisition:
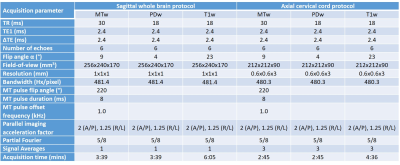
Seven healthy volunteers [2 females, age 32.9 ± 3.91 years (mean ± standard deviation (SD))] were scanned using a 3T Philips Achieva (Philips Healthcare, Best, The Netherlands), with a 32-channel head coil and multi-transmit technology. Both the brain and cord MPM protocols consisted of three (product) multi-echo 3D SPGR sequences (with bipolar gradients), with MT-(MTw), PD-(PDw) or T1-weighting (T1w) (details in Figure 1), and B1 mapping using actual flip angle imaging (AFI)8 (α=60°, TR1/TR2/TE=30/150/2.3ms).
Image Analysis:
Whole cord masks for each contrast acquired using both protocols were obtained semi-automatically using an active surface model9 implemented in Jim (www.xinapse.com). These segmentations were input when co-registering different contrasts to the mean PDw volume (using spinal cord toolbox (SCT)10). SCT was used to label cord vertebrae and generate binary cord masks, and to perform cord segmentation, giving white (WM) and grey matter (GM) binary masks.
Processing was performed using in-house written matlab code. R2* was estimated from the multi-echo PDw data, by linear fitting of the log of the signal. After averaging the within-contrast echoes, T1 and APD were calculated from the PDw and T1w images via a rational approximation of the SPGR signal3. The percentage MT saturation due to a single MT pulse (MTsat) was calculated from the PDw and MTw images by inserting the estimated APD and T1 values in the approximate MT-SPGR signal equation4,6. T1 maps were corrected for variations in the transmit field B1.
Binary cord masks were applied to maps to give mean cervical cord (C1-C5) parameter values obtained using each protocol, as well as GM and WM values for the cervical cord protocol. Paired t-tests were performed to test for differences in mean whole cord parameter values between protocols.
Results
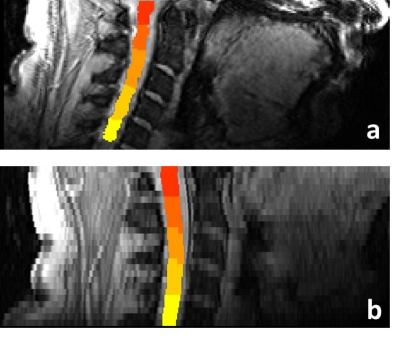
Example sagittal images with cord masks (levels C1-C5) overlaid for both protocols are shown in Figure 2.
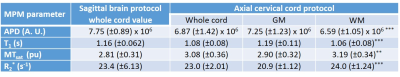
Mean parameter values measured in the cord using both protocols are given in Figure 3, in addition to GM and WM values obtained using the cord protocol. When averaged across all the subjects for each protocol, the maximum whole cord inter-subject CoV was 26% for R2* measured using the whole brain protocol, but was in the range of 5-12% for all other parameters measured using both protocols, consistent with literature7.
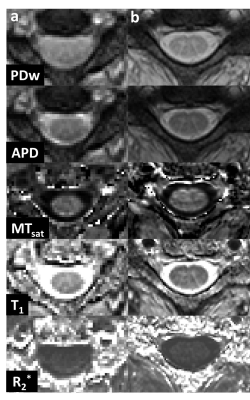
Single slice parameter maps of the same subject obtained using both protocols at cord level C3 are shown in Figure 4.
Paired t-tests showed no significant differences between whole cord parameters acquired using either protocol, except for APD (p=0.047).
Discussion
Cervical cord parameter values measured at 3T in healthy volunteers using a whole brain and dedicated cord protocol were consistent with literature values7,11,12, demonstrating the accuracy of both protocols, developed using only product sequences. With the exception of APD, mean whole cord parameters obtained from either protocol are within 10% of each other.
Paired t-tests showed no significant differences between parameters obtained from either protocol, except APD, which we attribute to the lack of calibration applied here (in the brain APD values are calibrated to the mean WM APD, however this was not possible since brain data were only acquired in one of the protocols).
Conclusions
It is possible to obtain several quantitative cervical cord parameters in a clinically feasible time (~17/12mins for brain/cord protocols) using both a protocol set up for whole brain MPM, and a dedicated cervical cord protocol. Inter-subject variation is low using either protocol, and both protocols give similar whole cord parameter values. Using the dedicated cord approach, it is feasible to perform accurate GM segmentation, giving tissue-specific measurements in addition to whole cord values. Improvements in image analysis and segmentation methods would be necessary to fully exploit cord data acquired using the brain protocol12.Acknowledgements
This study was funded by INSPIRED (a spinal cord imaging grant funded by the International Spinal Research Trust, Wings for Life, and CHNF). Funding was also received from the MS Society of the UK, and the Department of Health’s NIHR Biomedical Research Centres funding scheme. Additionally, this project has received funding from the European Union's Horizon 2020 research and innovation programme under the grant agreement No 681094, and is supported by the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract number 15.0137. We would also like to thank all the participants of the study.References
[1] Wheeler-Kingshott, CAM et al Neuroimage 84:1082-93 (2014);
[2] Weiskopf N, Helms G Proceedings ISMRM 16: 2241 (2008);
[3] Helms G et al Magnetic Resonance in Medicine 59:667-72 (2008);
[4] Helms G et al Magnetic Resonance in Medicine 60: 1396-1407 (2008);
[5] Helms G et al Magnetic Resonance in Medicine 64: 177-85 (2010);
[6] Weiskopf N et al Frontiers of Neuroscience7: 95 (2013);
[7] Samson RS et al. NMR in Biomedicine 26(12):1823-30 (2013);
[8] Yarnykh V et al Magnetic Resonance in Medicine 57(1):192-200 (2007) ;
[9] Horsfield MA et al. NeuroImage 50: 446-455 (2010);
[10] De Leener B et al NeuroImage 15:145(Pt A):24-43 (2017);
[11] Smith SA et al Magnetic Resonance in Medicine 60(1): 213–219 (2008);
[12] Battiston M et al Magnetic Resonance in Medicine; in press, published online 24th July 2017;
[12] Prados F et al NeuroImage 152 :312-329 (2017).
Figures