0664
Motion artifact quantification and localization for whole-body MRI1Section on Experimental Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 2Institute of Signal Processing and System Theory, University of Stuttgart, Stuttgart, Germany, 3Department of Radiology, University Hospital of Tuebingen, Tuebingen, Germany
Synopsis
Motion is still one of the major extrinsic factors degrading image quality. Automated detection of these artifacts is of interest, (i) if suitable prospective or retrospective correction techniques are not available/applicable, (ii) if human experts who judge the achieved quality are not present, or (iii) if a manual quality analysis of large databases from epidemiological cohort studies is impracticable. A convolutional neural network assesses and localizes the motion artifacts. This work extends the previously published method by proposing a general architecture for a whole-body scenario with varying contrast weightings. High accuracies of >90% were achieved in a volunteer study.
Introduction
MRI offers a broad variety of imaging applications with targeted and flexible sequence and reconstruction parametrization. Varying acquisition conditions and long examination times make MRI susceptible to imaging artifacts, amongst which motion is one of the major extrinsic factors deteriorating image quality.
Images recorded for clinical diagnosis are often inspected by a human specialist to determine the achieved image quality which can be a time-demanding and cost-intensive process. If insufficient quality is determined too late, an additional examination may be even required decreasing patient comfort and throughput. Moreover, in the context of large epidemiological cohort studies such as UK Biobank1 or German National Cohort2 the amount and complexity exceed practicability for a manual image quality analysis. Thus, in order to guarantee high data quality, arising artifacts need to be detected as early as possible to seize appropriate countermeasures, as e.g. prospective3-5 or retrospective correction techniques6-9. However, these methods are not always available and/or applicable. Moreover, when a human expert is not present or for large cohort studies, the potential presence of motion artifacts demands an automatic processing for a prospective quality assurance or retrospective quality assessment.
Previously proposed approaches for automated medical image quality analysis require the existence of a reference image and/or were only focused on specific sequences and scenarios10-12. Reference-free approaches13-17 are mainly metric-based driven to evaluate the quality on a coarse level.
In none of the previous approaches a motion artifact localization and quantification was conducted. We therefore proposed an automatic and reference-free motion artifact detection by a machine-learning convolutional neural network (CNN) approach18,19. In our previous study, the architecture was kept intentionally shallow because of the limited amount of data and investigations focused only on motion in head and abdominal region for one imaging sequence. In this work, we investigate a general architecture for motion artifact detection in a whole-body scenario with two contrast weightings.
Material and Methods
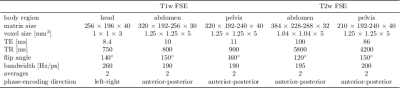
MR images were acquired on a 3T PET/MR (Biograph mMR, Siemens) from 18 healthy volunteers (3 female, 25±8y) with a T1w and T2w FSE sequence. The acquisition parameters for the respective body regions (head, abdomen, pelvis) are depicted in Tab.1. Each of the five sequences was acquired twice for every volunteer. During the first acquisition volunteers were asked to avoid movements (head, pelvis) or an end-expiratory breath-hold (T1w) respectively navigator-triggering (T2w) was conducted in the abdominal region. In the second acquisition, volunteers were asked to move their heads, hips (rigid deformation) or to breathe normal (non-rigid deformation). Images are normalized into an intensity range of 0 to 1 and partitioned into 50% overlapping patches of size 40x40x10 (APxLRxSI).
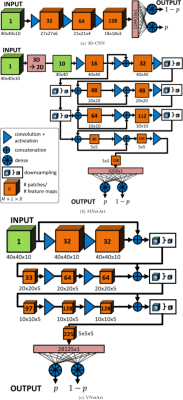
The proposed CNN architectures are depicted in Fig.2 and compared against the previously published 2D-CNN18 to output artifact probabilities p. 3D-CNN depicts the 3D extension of the 2D-CNN with three convolutional layers of N filter kernels/channels of size MxLxB with rectified linear unit (ReLU) activation, followed by a fully-connected dense layer with softmax decision.
The MNetArt is inspired by MNet20 consisting of four stages followed by a dense output layer. Each stage contains two convolutional layers with ReLU activation, an intermittent and finalizing concatenation layer. This resembles a residual path to forward feature maps in and between stages. The first three layers exhibit an additional max-pooling downsampling.
The VNetArt is inspired by VNet21 consisting of three stages followed by a dense output layer. Each stage has two convolutional layers with ReLU activation, a finalizing concatenation layer (residual path) and max-pooling downsampling.
The four architectures (2D-CNN, 3D-CNN, MNetArt, VNetArt) are trained by a leave-one-subject-out cross-validation on (i) the complete training database, (ii) the subset consisting only of T1w or T2w images, (iii) subsets leaving-out one body region. Categorical cross-entropy is minimized for given learning rate, $$$\ell_2$$$ regularization and dropout. Parameter ranges are estimated by the Baum-Haussler rule22 with a grid-search optimization. Testing was performed on the left-out subject to determine accuracy, sensitivity (true positive rate; TPR) and specificity (true negative rate; TNR).
Results and Discussion
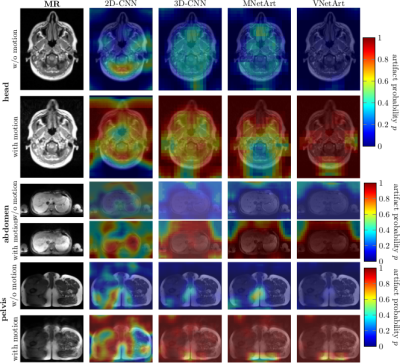
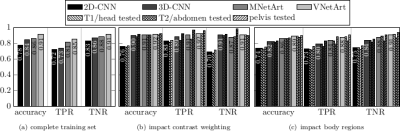
Fig.3 depicts exemplary subject slices overlaid with the derived and localized motion artifact probabilities. For this small moving subject a high accuracy of 98%/83%/90% in the head, abdomen and pelvis was achieved, respectively. Fig.4 illustrates superior performance of 3D processing and inclusion of residual paths. Performance is influenced by trained body regions (type of motion), but is independent of contrast weighting. Overall, VNetArt performs best for all metrics.Conclusion
Proposed architectures improve previously reported network by 17%. Automatic motion artifact quantification and localization is feasible in a whole-body setting of various imaging sequences with accuracies >90%.Acknowledgements
No acknowledgement found.References
[1] W. Ollier, T. Sprosen, and T. Peakman, “UK Biobank: from concept to reality,” Pharmacogenomics, vol. 6, no. 6, pp. 639-646, 2005. [2] F. Bamberg, H. U. Kauczor, S. Weckbach, C. L. Schlett, M. Forsting, S. C. Ladd, K. H. Greiser, M. A. Weber, J. Schulz-Menger, T. Niendorf, T. Pischon, S. Caspers, K. Amunts, K. Berger, R. Bulow, N. Hosten, K. Hegenscheid, T. Kroncke, J. Linseisen, M. Gunther, J. G. Hirsch, A. Kohn, T. Hendel, H. E. Wichmann, B. Schmidt, K. H. Jockel, W. Hoffmann, R. Kaaks, M. F. Reiser, and H. Volzke, “Whole-Body MR Imaging in the German National Cohort: Rationale, Design, and Technical Background,” Radiology, vol. 277, no. 1, pp. 206–20, 2015. [3] M. Zaitsev, J. Maclaren, and M. Herbst, “Motion artifacts in mri: A complex problem with many partial solutions,” J. Magn. Reson. Imaging., vol. 42, no. 4, pp. 887–901, 2015. [4] J. Maclaren, M. Herbst, O. Speck, and M. Zaitsev, “Prospective motion correction in brain imaging: a review,” Magn. Reson. Med., vol. 69, no. 3, pp. 621–36, 2013. [5] F. Godenschweger, U. Kagebein, D. Stucht, U. Yarach, A. Sciarra, R. Yakupov, F. Lusebrink, P. Schulze, and O. Speck, “Motion correction in mri of the brain,” Phys. Med. Biol., vol. 61, no. 5, pp. R32–56, 2016. [6] L. Feng, R. Grimm, K. T. Block, H. Chandarana, S. Kim, J. Xu, L. Axel, D. K. Sodickson, and R. Otazo, “Golden-angle radial sparse parallel mri: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric mri,” Magn. Reson. Med., vol. 72, no. 3, pp. 707–17, 2014. [7] J. Y. Cheng, T. Zhang, N. Ruangwattanapaisarn, M. T. Alley, M. Uecker, J. M. Pauly, M. Lustig, and S. S. Vasanawala, “Free-breathing pediatric mri with nonrigid motion correction and acceleration,” J. Magn. Reson. Imaging., vol. 42, no. 2, pp. 407–20, 2015. [8] G. Cruz, D. Atkinson, C. Buerger, T. Schaeffter, and C. Prieto, “Accelerated motion corrected three-dimensional abdominal mri using total variation regularized sense reconstruction,” Magn. Reson. Med., vol. 75, no. 4, pp. 1484–98, 2016. [9] T. Küstner, C. Würslin, M. Schwartz, P. Martirosian, S. Gatidis, C. Brendle, F. Seith, F. Schick, N.F. Schwenzer, B. Yang, and H. Schmidt, “Self-navigated 4d cartesian imaging of periodic motion in the body trunk using partial k-space compressed sensing,” Magn. Reson. Med., vol. 78, no. 2, pp. 632–644, 2017. [10] J. Oh, S. I. Woolley, T. N. Arvanitis, and J. N. Townend, “A multistage perceptual quality assessment for compressed digital angiogram images,” IEEE Trans. Med. Imaging., vol. 20, no. 12, pp. 1352–61, 2001. [11] J. Miao, D. Huo, and D. L. Wilson, “Quantitative image quality evaluation of MR images using perceptual difference models,” Med. Phys., vol. 35, no. 6, pp. 2541–53, 2008. [12] A. Ouled Zaid and B. B. Fradj, “Coronary angiogram video compression for remote browsing and archiving applications,” Comput. Med. Imaging. Graph., vol. 34, no. 8, pp. 632–41, 2010. [13] B. Mortamet, M. A. Bernstein, Jr. Jack, C. R., J. L. Gunter, C. Ward, P. J. Britson, R. Meuli, J. P. Thiran, and G. Krueger, “Automatic quality assessment in structural brain magnetic resonance imaging,” Magn. Reson. Med., vol. 62, no. 2, pp. 365–72, 2009. [14] J. P. Woodard and M. P. Carley-Spencer, “No-reference image quality metrics for structural MRI,” Neuroinformatics, vol. 4, no. 3, pp. 243–62, 2006. [15] M. D. Tisdall and M. S. Atkins, “Using human and model performance to compare MRI reconstructions,” IEEE Trans. Med. Imaging., vol. 25, no. 11, pp. 1510–7, 2006. [16] D. Atkinson, D. L. Hill, P. N. Stoyle, P. E. Summers, and S. F. Keevil, “Automatic correction of motion artifacts in magnetic resonance images using an entropy focus criterion,” IEEE Trans. Med. Imaging, vol. 16, no. 6, pp. 903–10, 1997. [17] K. P. McGee, A. Manduca, J. P. Felmlee, S. J. Riederer, and R. L. Ehman, “Image metric-based correction (autocorrection) of motion effects: analysis of image metrics,” J. Magn. Reson. Imaging, vol. 11, no. 2, pp. 174–181, 2000. [18] T. Küstner, A. Liebgott, L. Mauch, P. Martirosian, F. Bamberg, K. Nikolaou, B. Yang, F. Schick, and S. Gatidis, “Automated reference-free detection of motion artifacts in magnetic resonance images,” Magn. Reson. Mater. Phys., Biol. Med., Sep 2017. [19] T. Küstner, A. Liebgott, L. Mauch, P. Martirosian, K. Nikolaou, F. Schick, B. Yang, and S. Gatidis, „Automatic reference-free detection and quantification of MR image artifacts in human examinations due to motion,” ISMRM Proceedings, p. 1278, 2017. [20] R. Mehta and J. Sivaswamy, “M-net: A Convolutional Neural Network for deep brain structure segmentation,” in IEEE International Symposium on Biomedical Imaging (ISBI), April 2017, pp. 437–440. [21] F. Milletari, N. Navab, and S.-A. Ahmadi, “V-Net: Fully Convolutional Neural Networks for volumetric Medical Image Segmentation,” ArXiv e-prints, June 2016. [22] E.B. Baum and D. Haussler, “What Size Net Gives Valid Generalization?,” Neural Comput., vol. 1, no. 1, pp. 151–160, Mar. 1989.Figures