0659
Combining Multi-Site/Study MRI Data: A Novel Linked-ICA Denoising Method for Removing Scanner and Site Variability from Multi-Modal MRI Data1Imaging Center, Harvard Medical School, McLean Hospital, Belmont, MA, United States, 2Dalian University of Technology, Dalian, China, 3Oxford University, Oxford, United Kingdom, 4Harvard Medical School, Beth Israel Deaconess Medical Center, Boston, MA, United States, 5University of Arizona, Tucson, AZ, United States
Synopsis
Large multi-site studies that pool magnetic resonance imaging (MRI) data across research sites present exceptional opportunities to advance neuroscience and enhance reproducibility of neuroimaging research. However, inconsistent MRI data collection platforms and scanning sequences both introduce systematic variability that can confound the true effect of interest and make the interpretation of results obtained from combined data difficult. Unfortunately, methods to address this problem are scant. In this study, we propose a novel denoising approach for multi-site, multi-modal MRI data that implements a data-driven linked independent component analysis to efficiently identify scanner/site-related effects for removal.
Introduction
Large multi-site studies that pool magnetic resonance imaging (MRI) data across research sites present exceptional opportunities to advance neuroscience and enhance reproducibility of neuroimaging research1,2. Currently, there are more than a dozen ongoing large-scale multi-site neuroimaging studies (e.g., NIH-funded studies and UK Biobank). The strength of these large-scale studies lies in combining multi-site data to create large datasets that overcome limitations of small neuroimaging studies. However, both scanner and site variability are confounds that hinder pooling data collected across different sites or across different scanner software on the same hardware, even when all acquisition protocols are harmonized3,4. These confounds degrade statistical analyses, leading to incorrect or spurious findings. Unfortunately, methods to address this problem are scant. We propose a novel denoising approach for multi-site, multi-modal MRI data that implements a data-driven linked independent component analysis (LICA)5,6 to efficiently identify scanner/site-related confounds for removal. Removing these confounds results in denoised data that can be combined across studies to improve modality-specific statistical processing.
Methods
Data: Data from 133 subjects (62 chronic heavy marijuana smokers and 71 healthy controls (HC)) from 6 different studies were used. All data were collected using the same Siemens 3T Trio, but with 3 different scanner software versions (SSWV), VA23A, VA25A and VB17A. VA23A and VA25A were used prior to a major hardware and software upgrade of the Trio (TIM upgrade), while VB17A was used post-TIM. Acquisition sequences also differed across the studies, thus, the main confounds for combining data were SSWV and STUDY variability.
Data processing: Modality-specific preprocessing pipelines were used to produce outcome images for each participant, including: modulated grey matter (GM) images generated by FSL-VBM and vertex-wise cortical thickness (CT) and pial surface area (PSA) maps estimated by FreeSurfer, fractional anisotropy (FA), mean diffusivity (MD) and tensor mode (MO) images calculated using FSL FDT, and brain activation maps estimated by FSL FEAT analysis of functional MRI (fMRI) data collected during the Multi Source Interference Task. For each modality, a “subject” series was created by normalizing all images to MNI152 space, then concatenating across all participants into a single data file.
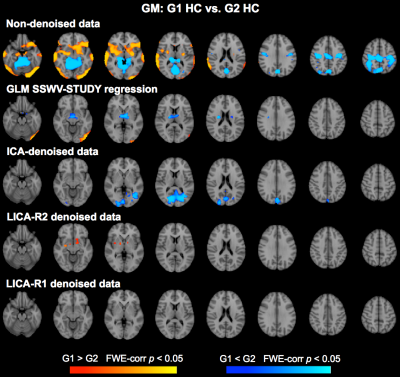
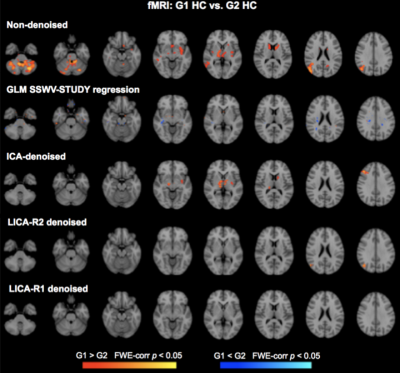
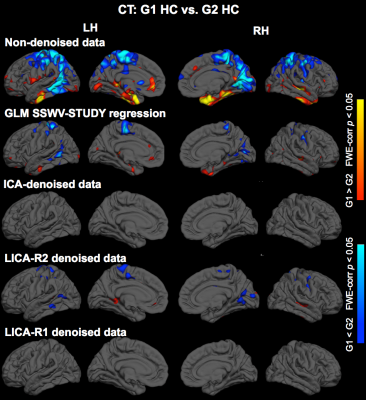
Denoising: Subject-series for all 7 modalities were analyzed simultaneously using LICA to derive 15 multi-modal spatial components. Subject-loadings (SL) for each component were assessed for relationships with SSWV, STUDY and participant variables using linear regression; those with SL that related only with SSWV and STUDY were identified for denoising. Two approaches for LICA-denoising were tested: LICA-R1, which applies a single multivariate regression (MVR) of the SL for all noise components against the participant-series for each modality to remove the noise effects, and LICA-R2, which uses a two-stage MVR to remove noise components by regressing the LICA spatial maps against each subject-series to obtain subject-specific regression weights that are then regressed against the subject-series to remove the noise effects. We compared the performance of LICA-R1/R2 with two other approaches for addressing scanner confounds when combining MRI data across studies/sites: a higher-level GLM with a site/study covariate (SSC-GLM) included in the group-level model, and modality-specific ICA denoising based on FSL MELODIC7. While all data were used to conduct the LICA to identify noise components, we constructed test data for each modality by splitting the data from HC into two “groups”, defined based on SSWV and STUDY variables. Thus any observed differences when comparing the two groups can be attributed to differences introduced by SSWV or STUDY. Group differences in each modality were assessed before and after denoising, using two-group t-tests with non-parametric permutation testing in FSL’s Randomise with 5000 permutations to achieve a significance level of p < 0.05, corrected for family-wise error.
Results
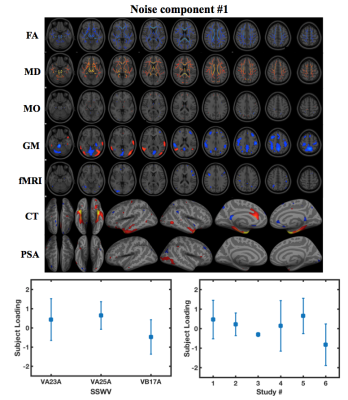
Three noise components identified from 15 LICA components were used for LICA-R1/R2 denoising. The first revealed global effects in FA and MD and region-specific effects in GM, fMRI, CT and PSA (Fig. 1). The second revealed region-specific effects in FA, MD, GM, CT and PSA, while the third revealed effects in GM. Comparison of LICA-R1/R2 with SSC-GLM and modality-specific ICA based on the denoising performance on GM, fMRI and CT data (Figs. 2, 3 and 4), shows that SSC-GLM and ICA-based denoising were modestly effective at removing confounds. LICA-R1 showed superior performance over all methods in denoising scanner effects, removing them completely for each modality.
Discussion and Conclusion
A new method for denoising is proposed for removing site/scanner effects from multi-site/study MRI data. The proposed method (LICA-R1) is superior compared to existing strategies we tested and has great potential for large-scale multi-site studies to produce combined data free from study/site confounds.
Acknowledgements
This project was funded by NIDA R01 DA037265.References
1. Van Horn JD, Toga AW. Human Neuroimaging as a “Big Data” Science. Brain Imaging Behav. 2014; 8: 323–331.
2. Varoquaux G. Cross-validation failure: Small sample sizes lead to large error bars. NeuroImage. 2017; PMID: 28655633.
3. Jovicich J, Marizzoni M, Sala-Llonch R, et al. Brain morphometry reproducibility in multi-center 3T MRI studies: A comparison of cross-sectional and longitudinal segmentations. Neuroimage. Elsevier Inc. 2013; 83: 472-484.
4. Venkatraman VK, Gonzalez CE, Landman B, et al. Region of interest correction factors improve reliability of diffusion imaging measures within and across scanners and field strengths. NeuroImage. 2015; 119: 406-416.
5. Groves AR, Beckmann CF, Smith SM, et al. Linked independent component analysis for multimodal data fusion. NeuroImage. 2011; 54: 2198-2217.
6. Groves AR, Smith SM, Fjell AM, et al. Benefits of multi-modal fusion analysis on a large-scale dataset: life-span patterns of inter-subject variability in cortical morphometry and white matter microstructure. NeuroImage. 2012; 63: 365-380.
7. Chen J, Liu J, Calhoun VD, et al. Exploration of scanning effects in multi-site structural MRI studies. J Neurosci. Methods. 2014; 230: 37-50.
Figures