0656
Construction of Spatiotemporal Infant Cortical Surface Atlas of Rhesus Macaque1Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
Rhesus macaque is a widely used animal model helping understand neural development in the human brain. Available adult macaque brain atlases are not suitable for infant studies, due to their dramatic brain difference. Building age-matched atlases is thus highly desirable yet still lacking. In this study, using MRI scans for 32 healthy rhesus monkeys, we constructed the first spatiotemporal cortical surface atlas for macaques aged from 2 weeks to 24 months, which were further equipped with developmental-trajectory-based parcellation maps. These surface atlases and parcellation maps will greatly facilitate the early brain development studies of macaques.
Introduction
Studying the rhesus brain development based on Magnetic Resonance (MR) images is important not only for understanding the maturation of normal brains, but also for investigating the intervention and treatment of neurodevelopmental disorders1,2. However, available adult monkey brain atlases are not suitable for the studies during the early postnatal stage. This is because the MRI of infant rhesus brains have dramatic changes in image contrast, intensity, appearance and folding degree across different ages. Therefore, for the first time, we developed the spatiotemporal (4D) cortical surface atlas of infant rhesus macaques to comprehensively characterize the early postnatal brain development. Specifically, our 4D macaque atlas was created at 13 time-points, from 2 weeks to 24 months of age, using 138 serial MRI scanned for 32 rhesus monkeys. In addition, we further parcellated our 4D atlas into distinct regions purely based on the cortical developmental trajectories. Our parcellation results can reflect the underlying cortical cytoarchitectonic changes and thus help define the microstructurally distinctive cortical regions.Methods
This study was performed based on a public rhesus macaque neurodevelopment dataset with 32 rhesus monkeys, in which each monkey has 4 to 5 longitudinal MRI scans during early postnatal stages with acquisition parameter detailed in1. We applied our in-house developed tools for infant brain tissue segmentation and cortical surface reconstruction3-5. All cortical surfaces were then mapped onto a standard sphere to facilitate atlas construction and analysis, with each vertex attached with cortical morphological features (e.g., cortical thickness, sulcal depth, average convexity, and curvature). Directly building the 4D cortical surface atlas using group-wise surface registration to align all subjects at different ages separately would result in temporally-inconsistent atlases, which may influence the analysis of early brain development. To tackle this challenge and leverage the within-subject longitudinal constraints, an advanced registration strategy4 was adopted in this study. Specifically, we first performed the intra-subject registration to obtain within-subject mean cortical surfaces. Then, we performed the inter-subject registration of within-subject mean surfaces to obtain unbiased and longitudinally-consistent 4D cortical surface atlases. Based on the established longitudinal and cross-sectional cortical correspondences, the developmental trajectory was defined at vertex-level by concatenating the corresponding cortical attributes across different time-points. The Pearson’s correlation was computed between any two vertices, forming a similarity matrix for each subject. This matrix was then normalized to [0, 1] and averaged across different subjects. We then applied the spectral clustering method6 on the averaged similarity matrix to perform the parcellation. We used the local surface area in this study for atlas parcellation, which, however, can be easily replaced by other cortical attributes, e.g., cortical thickness and cortical folding.Results
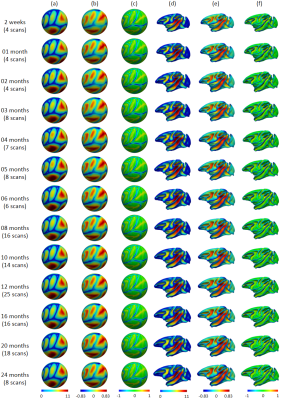
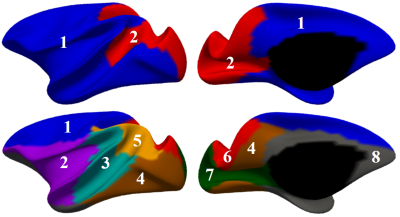
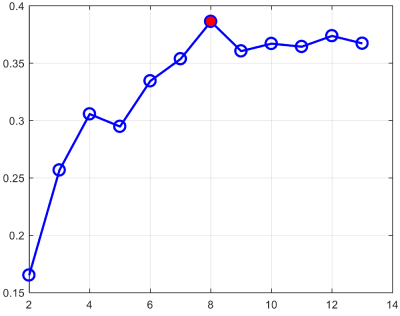
Fig. 1 shows our constructed 4D cortical surface atlas of rhesus macaques during the first 24 months, including 13 time-points at 2 weeks, 1, 2, 3, 4, 5, 6, 8, 10, 12, 16, 20 and 24 months of age. We can observe that major cortical folds are well established since birth and remain relatively stable during brain development. However, both average convexity and sulcal depth develop rapidly during the first 6 months, and then change gradually. These observations indicate the necessity of building an age-specific atlas, especially for early postnatal stage undergoing dynamic development. To capture the main pattern in developmental regionalization of the cortical structure, we first show the two-cluster parcellation map (top row of Fig. 2), which identifies a clear anterior-posterior division. To determine an appropriate number of clusters, we computed the widely used silhouette coefficient7. As shown in Fig. 3, according to the peak of the silhouette coefficients, we identified 8 clusters and presented the parcellation map in the bottom row of Fig. 2, from which we can observe that the obtained clusters are consistent with structurally and functionally meaningful regions. According to the labeled numbers in the figure, these regions approximately correspond to: (1) superior and middle frontal, (2) inferior frontal, insula, and temporal pole, (3) superior temporal, (4) inferior temporal and precuneus, (5) supra-marginal, (6) superior parietal, (7) occipital, and (8) limbic and cingulate. This parcellation map shows meaningful correspondences to existing neuroscience knowledge.Conclusion
We have built the first 4D cortical surface atlas for infant rhesus monkeys aged from 2 weeks to 24 months, with dense time points and in an unbiased and longitudinally-consistent manner. We have also unprecedentedly parcellated our 4D cortical surface atlas into distinct regions using developmental trajectories of local surface area. Our 4D atlas equipped with development-based parcellation maps will be a good reference for visualization, spatial normalization, analysis and comparison among various studies on both normal and abnormal monkey brain development.Acknowledgements
This work was partially supported by NIH grants (MH108914 and MH107815). We thank Dr. Matin A. Styner and his co-authors for making the UNC-Wisconsin Neurodevelopment Rhesus MRI Database publicly available.References
1. Young JT, Shi Y, Niethammer M, et al. The UNC-Wisconsin Rhesus Macaque Neurodevelopment Database: A Structural MRI and DTI Database of Early Postnatal Development. Front Neurosci. 2017;11.
2. Scott JA, Grayson D, Fletcher E, et al. Longitudinal analysis of the developing rhesus monkey brain using magnetic resonance imaging: birth to adulthood. Brain Struct Funct. 2016;221(5):2847-2871.
3. Li G, Nie J, Wang L, et al. Measuring the dynamic longitudinal cortex development in infants by reconstruction of temporally consistent cortical surfaces. Neuroimage. 2014;90:266-279.
4. Li G, Wang L, Shi F, Gilmore JH, Lin W, Shen D. Construction of 4D high-definition cortical surface atlases of infants: Methods and applications. Med Image Anal. 2015;25(1):22-36.
5. Wang L, Gao Y, Shi F, et al. LINKS: Learning-based multi-source IntegratioN frameworK for Segmentation of infant brain images. NeuroImage. 2015;108:160-172.
6. Ng AY, Jordan MI, Weiss Y. On spectral clustering: Analysis and an algorithm. Advances in neural information processing systems; 2002.
7. Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53-65.
Figures