0655
Accelerated high b-value diffusion-weighted MRI for higher-order diffusion analysis using a phase-constrained low-rank tensor model1Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States, 2Radiation Oncology, University of Michigan, Ann Arbor, MI, United States
Synopsis
DWI acquired with b-values greater than 1000 s/mm2 and higher-order diffusion analyses based on such DWI series have the potential to improve tumor differentiation, while the extended sampling of b-values makes the acquisition time inconveniently long. We propose an acceleration scheme that sparsely samples k-space and reconstructs images using a new low-rank tensor model which exploits both global and local low-rank structure. Under an acceleration factor of 8, parameter mapping results on one simulated and 7 patient datasets show improved accuracy over another low-rank tensor model that exploits global correlation only, and comparable accuracy to clinically used four-fold GRAPPA reconstruction.
Introduction
Diffusion-weighted MR images (DWI) acquired with b-values greater than 1000 s/mm2 and higher-order diffusion analyses based on high b-value DWI series have the potential to improve tumor differentiation1-3. However, extended sampling of b-values makes the acquisition time inconveniently long for clinical use. We propose to reduce scan time by using sparse k-space sampling and a new low-rank tensor model for DWI reconstruction which exploits both global and local low-rank structure of DWI data, and integrates partial Fourier acquisition naturally.Method
The target DWI data is acquired using an imaging matrix of $$$N_{x}\times N_{y}$$$, with $$$N_{c}$$$ coils and $$$N_{b}$$$ b-values. To model the local low-rank structure (i.e., correlation between neighboring k-space samples), we first organize k-space data at each b-value into a block-Hankel matrix $$$H_{b}\in\mathbb{C}^{N_{c}\cdot w^2 \times (N_{x}-w+1)(N_{y}-w+1)}$$$ using the SAKE4 method with a window size $$$w$$$. Next we stack block-Hankel matrices at different b-values along the third dimension to form a tensor $$$\mathcal{X} \in \mathbb{C}^{N_{c}\cdot w^2 \times (N_{x}-w+1)(N_{y}-w+1) \times N_{b}}$$$ to exploit the global low-rank structure (i.e., correlation of signal decays across voxels). We propose the following constrained image reconstruction scheme that enforces low-rankness of $$$\mathcal{X}$$$
$$\begin{align}\label{fcmo} &\hat{\mathbf{y}},\hat{\mathbf{x}},\hat{\mathcal{X}} = \textrm{arg min} \|\mathbf{d}-\Omega \mathbf{y}\|_{2}^{2}+\lambda \mathbf{R}(\mathcal{X}) \nonumber \\ & \textrm{s.t.}\quad \mathbf{y} = \mathcal{F}\mathbf{P}\mathbf{x},\quad \mathcal{X} = \mathcal{H}\mathcal{F}\mathbf{P_{1}}\mathbf{x},\quad \mathbf{x}\in \mathbb{R}^{N_{x}N_{y}N_{c}N_{b}},\end{align}$$
where $$$\mathbf{d}$$$ denotes the k-space samples, $$$\Omega$$$ is the sampling operator, $$$\mathbf{P}$$$ is the phase maps of each b-value/coil, estimated from the center of k-space, and $$$\mathbf{P_{1}}$$$ is the reference coil phase map estimated at b = 0 s/mm2 . By introducing $$$\mathbf{P}$$$ and $$$\mathbf{P_{1}}$$$, we seek to correct phase variations across b-values that would invalidate the low-rank assumption, while maintaining coil phases needed for multi-coil combination. Furthermore, by forcing $$$\mathbf{x}$$$ to be real, our method incorporates the POCS method for partial Fourier acquisition5. $$$\mathcal{H}$$$ is the block-Hankel matrix operator. We chose the regularizer $$$\mathbf{R}$$$ as a hard constraint on the tensor $$$n$$$-rank6 of $$$\mathcal{X}$$$ such that $$$(\textrm{rank}(\mathbf{X}_{(1)}),\textrm{rank}(\mathbf{X}_{(2)}),\textrm{rank}(\mathbf{X}_{(3)})) \leq (r_1,r_2,r_3)$$$, where $$$\mathbf{X}_{(i)}$$$ is the $$$i$$$th order matrix unfolding of $$$\mathcal{X}$$$ . We solve the problem using an ADMM algorithm where the augmented Lagrangian function is written as
$$ L(\mathbf{x},\mathbf{y},\mathbf{u}_{1},\mathbf{u}_{2},\mathcal{X})=\|\mathbf{d}-\Omega \mathbf{y}\|_{2}^{2}+\lambda \mathbf{R}(\mathcal{X})+\mu_{1}(\|\mathbf{y}-\mathcal{F}\mathbf{P}\mathbf{x}+\mathbf{u}_{1}\|_{2}^{2}-\|\mathbf{u}_{1}\|_{2}^{2}) +\mu_{2}(\|\textrm{vec}(\mathcal{X}-\mathcal{H}\mathcal{F}\mathbf{P_{1}}\mathbf{x}+\mathbf{u}_{2})\|_{2}^{2}-\|\textrm{vec}(\mathbf{u}_{2})\|_{2}^{2}) $$
We solve the subproblem of $$$\mathcal{X}$$$ using truncated multi-linear singular value decomposition7. All other subproblems have closed-form solutions.
Evaluation
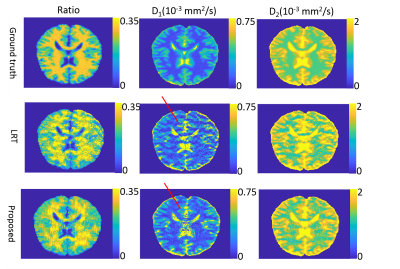
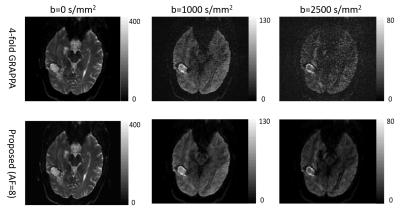
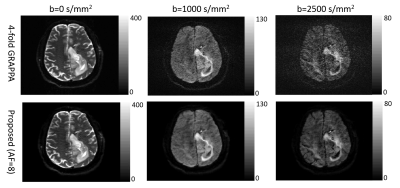
Under IRB approval, 7 patients with glioblastoma were scanned on a 3T scanner (SKYRA, Siemens Healthineers) with 20-channel coil arrays using a DW-EPI sequence with diffusion weighting in 3 orthogonal directions. Eleven b-values were sampled uniformly from 0 s/mm2 to 2500 s/mm2. Four-fold parallel imaging and partial Fourier were applied and the acceleration factor (AF) was 4.5. We further retrospectively undersampled the datasets along the phase-encoding direction only to achieve an AF of 8. The center of k-space was fully sampled and the peripheral k-space was randomly undersampled. A simulated DWI dataset was also generated using a brain phantom from brainweb8 and the same imaging parameters as the patient scan. Bi-exponential9 decays were simulated for brain tissues. Coil sensitivities and k-space noise were estimated from the patient data and linear phase variations across b-values were further added10. The simulated dataset was undersampled the same way as the patient dataset. Figure 1 shows the sampling scheme. Before performing constrained image reconstruction, we first calculated a GRAPPA11 kernel from the auto-calibration region at b = 0 s/mm2 to fill up the regularly undersampled k-space center for other b-values, and estimated phase maps from the center. We fitted two higher-order diffusion models to our reconstructed images: bi-exponential model9 for simulated data and stretched model1 for patient data. We compared our method to another low-rank tensor-based method (LRT method)12.Result
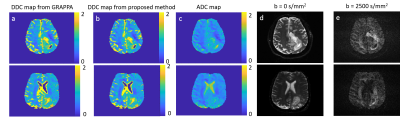
The root-mean-square error of our reconstruction on the simulated data is 3.1%, compared to the noise-free ground truth. Figure 2 shows parameter maps fitted by the bi-exponential model. Comparing to the LRT method, our method reduces the error by 18.9%, 39.6% and 7.1% for the three model parameters (two diffusion coefficients and one ratio). Figures 3 and 4 show example reconstructed patient images. No systematic difference is observed between our method with AF=8 and four-fold GRAPPA reconstruction, while our method improves the SNR at b>1000 s/mm2 by 20.1% across patients. Figure 5 shows the DDC maps fitted using the stretched model on GRAPPA as well as our reconstruction. The mean absolute difference of DDC between GRAPPA and our method is 0.02×10-3 mm2/s across patients.Discussion and Conclusion
A new low-rank tensor model that exploits both local and global low-rank structure achieves performance superior to models that exploit global low-rank structure only. Image reconstructions using undersampled data with AF= 8 can support higher-order diffusion analysis with accuracy comparable to parallel imaging-only reconstructions currently used clinically.Acknowledgements
This work is supported by NIHR01EB016079 and Rackham Barbour Scholarship.References
1. T.C. Kwee, C.J. Galbán, C. Tsien, L. Junck, P.C. Sundgren, M.K. Ivancevic, T.D. Johnson, C.R. Meyer, A. Rehemtulla and B.D. Ross, “Comparison of apparent diffusion coefficients and distributed diffusion coefficients in high-grade gliomas,” Journal of Magnetic Resonance Imaging, vol. 31, no. 3, pp. 531–537, 2010.
2. B.A. Hoff, T.L. Chenevert, M.S. Bhojani, T.C. Kwee, A. Rehemtulla, D. Le Bihan, B.D. Ross and C.J.Galbán, “Assessment of multi exponential diffusion features as MRI cancer therapy response metrics,” Magnetic resonance in medicine, vol. 64, no. 5, pp. 1499–1509, 2010.
3. P.P. Pramanik, H.A. Parmar, A.G. Mammoser, L.R.Junck, M.M. Kim, C. Tsien, T.S. Lawrence, and Y. Cao, “Hypercellularity components of glioblastoma identified by high b-value diffusion-weighted imaging,” International Journal of Radiation Oncology Biology Physics, vol. 92, no. 4, pp. 811–819, 2015.
4. P.J. Shin, P.E. Larson, M.A. Ohliger, M. Elad, J.M.Pauly, D.B. Vigneron, and M. Lustig, “Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion,” Magnetic resonance in medicine, vol. 72, no. 4, pp. 959–970, 2014.
5. E.M. Haacke, E.D. Lindskogj, and W. Lin, “A fast, iterative,partial-Fourier technique capable of local phasere covery,” Journal of Magnetic Resonance, vol. 92, no.1, pp. 126–145, 1991.
6. L. De Lathauwer, B. De Moor, and J. Vandewalle, “A multilinear singular value decomposition,” SIAM journal on Matrix Analysis and Applications, vol. 21, no. 4,pp. 1253–1278, 2000.
7. N. Vervliet, O. Debals, L. Sorber, M. Van Barel, and L. De Lathauwer, “Tensorlab 3.0,” Mar. 2016, Available online.
8. D.L. Collins, A.P. Zijdenbos, V. Kollokian, J.G. Sled,N.J. Kabani, C.J. Holmes, and A.C. Evans, “Design and construction of a realistic digital brain phantom,” IEEE transactions on medical imaging, vol. 17, no. 3,pp. 463–468, 1998.
9. R.V. Mulkern, H.P. Zengingonul, R.L. Robertson, P. Bogner, K.H. Zou, H. Gudbjartsson, C.R. Guttmann, D. Holtzman, W. Kyriakos, and F.A. Jolesz, “Multicomponent apparent diffusion coefficients in human brain: relationship to spin-lattice relaxation,” Magnetic resonance in medicine, vol. 44, no. 2, pp. 292–300,2000.
10. A.T. Van, D.C. Karampinos, J.G. Georgiadis, and B.P.Sutton, “k-space and image space combination for motion artifact correction in multicoil multishot diffusion weighted imaging,” in Proc. EMBC. IEEE, 2008, pp.1675–1678.
11. M.A. Griswold, P.M. Jakob, R.M. Heidemann, M. Nittka, V. Jellus, J. Wang, B. Kiefer, and A. Haase, “Generalized autocalibrating partially parallel acquisitions(GRAPPA),” Magnetic resonance in medicine, vol. 47,no. 6, pp. 1202–1210, 2002.
12. J.D. Trzasko and A. Manduca, “A unified tensor regression framework for calibrationless dynamic, multichannel MRI reconstruction,” in Proceedings of the 21st Annual Meeting of ISMRM, Salt Lake City, Utah, USA,2013, p. 603.
Figures