0654
A spatiotemporal fetal MRI atlas for multi-organ segmentation and growth analysis1School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom
Synopsis
We propose a comprehensive framework for spatiotemporal multi-organ fetal MR
Purpose
Longitudinal characterization of human organ development in-utero is very important for growth monitoring and anomaly detection. Significant fetal motion during MR image acquisition, especially around mid-gestation, makes automatic segmentation of fetal organs very challenging. Typically, manual segmentation is time-consuming and subjective. Therefore, an atlas at different gestational ages serves as an averaged anatomical structure and/or spatiotemporal segmentation prior is in high demand. Previous works1,2,3 mainly focused on brain atlases, while other important organs, such as lung, spine, and heart have only been rarely studied. It has been reported that the fetuses with a high risk of preterm born have smaller lung volumes compared to the healthy ones with similar gestational ages (GA)4. In this work, besides fetal brain, we study the fetal trunk area from the image reconstruction, registration and segmentation perspectives.Methods
Data: MR examinations for 78 volunteers, with uneventful pregnancies, ranging from 20.14 to 31.85 weeks GA, were performed on a 1.5 Tesla MR scanner. T2 weighted single shot Fast Spin Echo sequences were acquired along the subject’s axial, sagittal, and coronal orientations, with TR = 15000 ms, TE = (80 or 180) ms, filp angle α = 900, and voxel size = 1.25 x 1.25 x 2.5 mm. Atlas Construction: For each of the 78 subjects, 6-9 stacks of images were used to reconstruct a single volumetric fetal image using a slice-to-volume reconstruction (SVR) framework5,6, for an isotropic resolution of 0.75 mm. Brain and trunk were reconstructed separately to avoid the commonly existing non-linear movement in head and neck area. For each pair of images, we first affine registered them to each other for global motion correction. Then, free-form deformations (FFD) based non-linear registration7 were used to estimate the local deformations, which is given by a B-spline tensor product over the control point displacements. We used normalized mutual information as the similarity metric for the inter-subject registration. For each week, 15 subjects are used to generate an affine atlas template by averaging them in the transformed atlas space. We first use each image as a target and affine register it with each other subject; then average the transformation for each subject and transform it to the atlas space. The affine atlas at each GA week is acquired by averaging the transformations of the 15 selected subjects. To interpolate the intensity and shape variations between the subjects, we use a weighted averaging strategy similar to reference1 to compute the brain and trunk template, respectively. The initial affine atlas template can be constructed by the weighted average of each subject in the transformed space. Then we applied the FFD method to estimate the non-linear displacements between each subject and the intermediate affine template. Based on the combined affine and FFD parameters, each subject was transformed to the atlas space. Finally, the non-linear spatiotemporal atlas was constructed based on the non-rigid transformation of each subject and their kernel regression weights.
In this work, the annotated organs include the brain, lung, heart, and spine. 26 subjects with different GA were selected and manually labeled for lungs. The spine and heart were segmented using a combination of thresholding, morphology operations, and manual editing. The rest of brain and lung labels are acquired using the FFD based multi-atlas segmentation method8 based on 10-18 manual segmentations with closest GA.
Results and Discussion
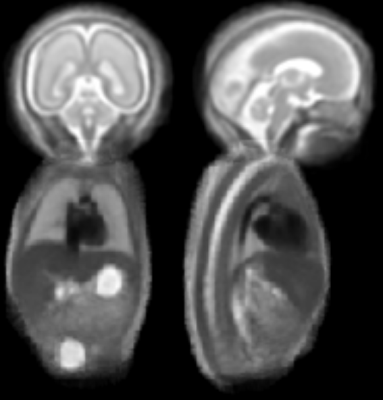
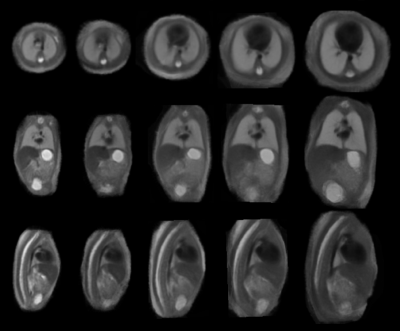
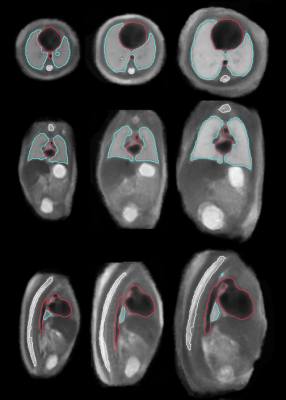
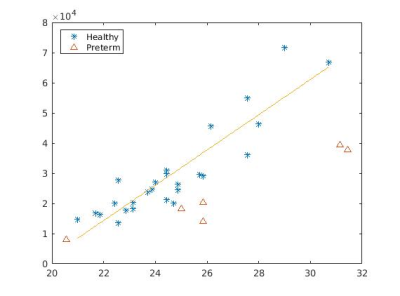
Figure 1 illustrates the constructed multi-organ atlas with combined brain and trunk view at 25 weeks GA. Figure 2 shows the spatiotemporal fetal trunk atlas at 22, 24, 26, 28, and 30 weeks GA. The atlas with annotations of lung, heart, and spine is shown in Figure 3, at 22, 26, and 30 weeks GA. We validated the multi-atlas segmentation method on lung volumes and the averaged dice coefficient value is 0.92. Figure 4 compares the lung volumes among 26 healthy and 6 high preterm risk foetuses4. It shows promising potential for using lung volumes for quantifying fetal growth and aid the diagnostic procedure for preterm subjects.
It is worth noting that the image contrast of the trunk atlas, especially in the heart and abdominal region, is generally superior to the reconstructed images. It would be averaging over several non-rigidly registered subjects that makes the atlas more homogeneous. This also indicates that the FFD method could potentially be used within the SVR framework to further improve the image quality.
Conclusion
We presented a framework for multi-organ spatiotemporal fetal atlasing and segmentation. The constructed atlas demonstrates that the proposed registration and weighting scheme can preserve the anatomical details and compensate for individual shape variabilities.Acknowledgements
This work was supported by the Wellcome Trust IEH Award [102431]; The authors acknowledge financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London and King’s College Hospital NHS Foundation TrustReferences
- Kuklisova-Murgasova, M., et al. A dynamic 4D probabilistic atlas of the developing brain. NeuroImage,2011. 54(4): p. 2750–2763
- Serag, A., et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. NeuroImage, 2012. 59: p.2255–2265
- Gholipour,A., et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Scientific Report,2017. 7(1):476.
- Story, L., et al. Magnetic resonance imaging assessment of lung volumes in fetuses at high risk of preterm birth. BJOG: An International Journal of Obstetrics and Gynaecology, 2017. 24:p.24-24
- Kuklisova-Murgasova, M., et al. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical Image Analysis, 2012. 16(8): p. 1550-1564.
- Kainz, B. et al. Fast volume reconstruction from motion corrupted stacks of 2D slices. Med Imaging, IEEE T, 2015. 34: p. 1901–1913
- Rueckert, D., et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE transactions on medical imaging, 1999. 18(8): p. 712-721.
- Heckemann, R. A., et al. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage, 2006. 33(1): p. 115–126
Figures