0635
COX-2 Alters the Metabolic Secretome in Triple Negative Human Breast Cancer XenograftsSantosh Kumar Bharti1, Paul T Winnard Jr.1, Yelena Mironchik1, Louis Dore-Savard2, Balaji Kirshnamachary1, and Zaver M Bhujwalla1,3
1Division of Cancer Imaging Research, Department of Radiology, Johns Hopkins University, School of Medicine, Baltimore, MD, United States, 2McGill University Health Centre and RI-MUHC, Montreal, QC, Canada, 3Department of Oncology, Johns Hopkins University, School of Medicine, Baltimore, MD, United States
Synopsis
Tumor interstitial fluid (TIF) contains the tumor secretome and forms a critical component of the tumor microenvironment. Cyclooxygenase-2 (COX2) mediates the inflammatory response of cells and is upregulated in cancers. In cancers, COX-2 expression has been related to increased invasion and metastasis. Here, for the first time, using 1H MR spectroscopy we characterized changes in the metabolic patterns of TIF in tumors derived from triple negative SUM-149 human breast cancer cells with COX-2 overexpressed. COX-2 overexpression significantly altered several fundamental metabolic pathways. These data provide new insights into the role of COX-2 in tumor aggressiveness, and identify new metabolic targets.
Introduction
Cyclooxygenase-2 (COX-2) is an active mediator of the inflammatory response of cells and plays an important role in the development, progression, invasion, and metastasis of several cancers including breast cancer [1-2]. Our ongoing studies of the tumor microenvironment (TME) of have focused on understanding and targeting COX-2 in triple negative breast cancer [3-4], the most lethal form of breast cancer. Tumor interstitial fluid (TIF), the milieu that contains the tumor secretome, is one of the least examined aspects of the TME because of the difficulty in sampling this fluid from tumors. Recently we adapted a previous published method [5] to sample TIF in human tumor xenografts in SCID mice [5]. Here, for the first time, we have sampled TIF from COX-2 overexpressing triple negative SUM-149 human breast cancer xenografts and empty vector SUM-149 xenografts. The metabolic profile of TIF from both tumor types was characterized using 1H magnetic resonance spectroscopy (1H MRS). These data represent the first ever characterization of the COX-2 metabolic secretome and reveal new insights into the role of COX-2 in altering the TME.Methods
The cloning, construction of a lentivirus vector expressing COX-2 gene, and the establishment of SUM-149 cells stably overexpressing COX-2 (SUM-COX-2) were reported by us previously [4]. Two million SUM-COX-2 or empty vector control cells (SUM-EV) were injected subcutaneously in female SCID mice to obtain tumor pieces for TIF chamber transplantation. A home-built TIF collection chamber [6] was inserted subcutaneously in female SCID mice with 4-6 1-2 mm tumor pieces packed around the chamber; the incision was closed with suture clips. Within 4-6 weeks the growing tumor encapsulated the chamber allowing the collection of TIF. Once tumors were ~ 400 mm3, TIF was carefully removed from the chamber in terminally anesthetized mice. TIF samples that were not clear fluid were discarded, resulting in 4 samples from SUM-COX-2 tumors and 4 samples from SUM-EV tumors. Each chamber yielded ~50mL of TIF that was diluted to a total volume of 500μL with D2O for high-resolution 1H MRS performed on an Avance III 750 MHz Bruker Spectrometer equipped with a 5 mm probe. 1H MR spectra with water suppression were acquired using a one-dimensional Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence with the following parameters: spectral width of 15495.8 Hz, time domain data points of 64K, effective 90° flip angle, relaxation delay 10s, acquisition time of 2.1s, 64 scans with 8 dummy scans, a receiver gain of 1030, and echo time 25ms. All spectra were processed using line broadening for exponential window function of 0.3 Hz prior to Fourier transformation, manually phased, and automatically baseline corrected using TOPSPIN 2.1. All surgical procedures and animal handling were performed in accordance with protocols approved by the Johns Hopkins University Institutional Animal Care and Use Committee, and conformed to the Guide for the Care and Use of Laboratory Animals published by the NIH. Immunoblot analysis of COX-2 expression was performed as previously described by us [4].Results and Discussion
SUM-COX-2 tumors showed consistently higher COX-2 expression compared to SUM-EV tumors (Figure 1). Representative spectra from SUM-COX-2 and SUM-EV tumors in Figure 2 demonstrate the profound differences in metabolites with COX-2 overexpression. These data are summarized for significant metabolic differences in the heat map in Figure 3. COX-2 overexpression resulted in a significant increase of lactate, glutamate, acetate, and succinate, and a significant decrease of glucose, glutamine, citrate, formate, and lipids; pyruvate tended to decrease. These data are displayed as quantitative bar graphs in Figure 4. The changes in lactate and lipids are consistent with our earlier observations where COX-2 downregulation in triple negative MDMB-231 human breast cancer cells resulted in a significant decrease of lactate and an increase of lipids and lipid droplets in intact perfused cells [7]. Here COX-2 overexpression increased glycolysis. Depletion of pyruvate observed here in COX-2 overexpressing cells would limit production of acetyl-CoA and consequently citrate to also diminish fatty acid synthesis/lipids. Increased succinate parallels the increase in glutamate/a-ketoglutarate, the upstream intermediate to succinate in the tricarboxylic acid cycle (TCA), and indicates an increased utilization/depletion of glutamine to supplement the TCA. Importantly, accumulation of succinate inhibits HIF-1a prolyl hydroxylase that stabilizes HIF-1a driven cancer promoting metabolic pathways such as enhanced glycolysis and increased ROS [8]. Moreover, increased succinate also, through the succinate/succinate dehydrogenase reaction, provides necessary electrons to the electron transport chain upstream of the COX-2 reaction [9]. The depletion of formate in COX-2 overexpressing cells indicates a disruption of several vital cellular metabolic pathways [9]. These data provide new insights into the role of COX-2 in the metabolic secretome and tumor metabolism, and identify metabolic pathways as potential targets for reducing the effects of COX-2 expression in cancer.Acknowledgements
This work was supported by NIH R35CA209960, R01CA82337 and NIH P30CA06973.References
- Wang D and Dubois RN. Eicosanoids and cancer. Nature reviews Cancer. 2010; 0(3):181-193.
- Singh-Ranger G, Salhab M and Mokbel K. The role of cyclooxygenase-2 in breast cancer: review. Breast Cancer Res Treat. 2008; 109(2):189-198.
- Stasinopoulos I, O'Brien DR, Wildes F, Glunde K and Bhujwalla ZM. Silencing of cyclooxygenase-2 inhibits metastasis and delays tumor onset of poorly differentiated metastatic breast cancer cells. Mol Cancer Res. 2007; 5(5):435-442.
- Krishnamachary B, Stasinopoulos I, Kakkad S, Penet MF, Jacob D, Wildes F, Mironchik Y, Pathak AP, Solaiyappan M, Bhujwalla ZM. Breast cancer cell cyclooxygenase-2 expression alters extracellular matrix structure and function and numbers of cancer associated fibroblasts. Oncotarget. 2017 Mar 14;8(11):17981-17994.
- Gullino PM, Clark SH, Grantham FH. The Interstitial Fluid of Solid Tumors. Cancer Res. 1964 Jun;24:780-94.
- Dore-Savard L, Lee E, Kakkad S, Popel AS, Bhujwalla ZM. The Angiogenic Secretome in VEGF overexpressing Breast Cancer Xenografts. Scientific reports. 2016;6:39460.
- Shah T, Stasinopoulos I, Wildes F, Kakkad S, Artemov D, Bhujwalla ZM. Noninvasive imaging identifies new roles for cyclooxygenase-2 in choline and lipid metabolism of human breast cancer cells. NMR in biomedicine. 2012;25(5):746-54.
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG Mansfield KD, Pa Y, Simon C, Thompson CB, Gottlieb E.Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-a prolyl hydroxylase. Cancer Cell. 2005;7:77-85.
- Mills E, O’Neill LAJ. Succinate: a metabolic signal in inflammation. Trends in Cell Biology. 2014;24(5):313-320.
- Morrow GP, MacMillan L, Lamarre SG, Young SK, MacFarlane AJ, Brosnan ME, Brosnan JT. In Vivo Kinetics of Formate Metabolism in Folate-deficient and Folate-replete Rats. 2015;290(4):2244-2250
Figures
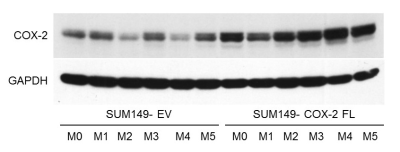
Figure
1: Immunoblot
of COX-2 expression in tumors derived from SUM-COX-2 (n=6) and SUM-EV (n=6) cells. GAPDH was used as a loading control. Higher COX-2 expression is consistently
observed in the SUM-COX-2 tumors.
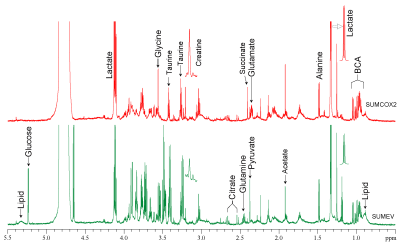
Figure
2: Representative
1H MR spectra of tumor interstitial fluid obtained from SUM-COX-2
(red) and SUM-EV (green) tumors.
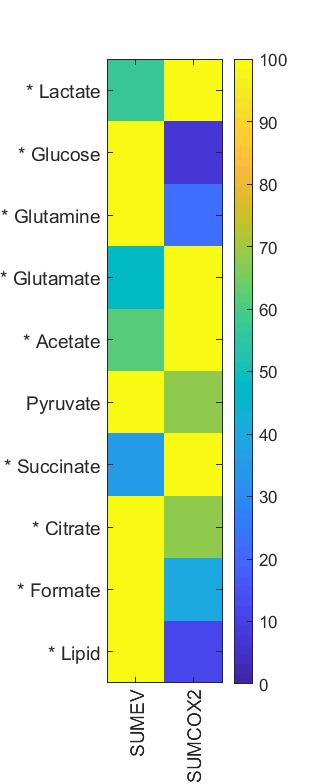
Figure
3: Heat
map representing significant differences in metabolites between TIF obtained
from SUM-EV (n=4) and SUM-COX-2 (n=4)
tumors. (* represents P<0.05, two
tailed t-test; pyruvate P<0.05 with one-tailed t-test)
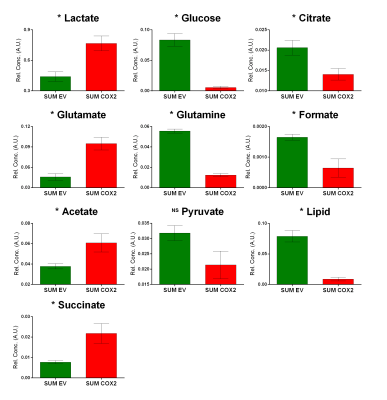
Figure 4: Quantitative
estimation of metabolites in TIF obtained from SUM-EV (n=4) and SUM- COX-2 (n=4)
tumors. Values represent mean±SE. Statistical significance was determined using an
unpaired two-tailed student-t test; * represents P<0.05: pyruvate P<0.05
with one-tailed t-test.