0598
Real-time acquisition, reconstruction, and mixed-reality display system for 2D and 3D cardiac MRI1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Department of Radiology, School of Medicine, Case Western Reserve University, Cleveland, OH, United States
Synopsis
Cardiac images suitable for 3D visualization are acquired, reconstructed, and displayed in real-time using the Microsoft HoloLens. This system could be used for guiding cardiac (or other) interventions, or be used to view time-resolved 2D or 3D datasets to facilitate the visualization of anatomical changes through time.
Introduction
The aim of this work was to develop an online real-time acquisition, reconstruction, and intuitive display system for 2D and 3D guidance of interventional procedures. Currently, MR guidance for cardiac interventions typically consists of either 2D real-time imaging with one or more slices, or display of 3D datasets collected pre-operatively. However, these methods either do not capture the full 3D environment, or they do not reflect the real-time positions of tissues and devices. An acquisition and reconstruction system for real-time, 3D cardiac imaging was recently proposed1, where each volume was displayed as a tiled set of 2D images, rather than in a 3D manner in which spatial relationships between partitions could be easily visualized. In this work, the highly undersampled data acquisition and fast implementation of through-time radial GRAPPA2 described previously are combined with a mixed-reality 3D display using the Microsoft HoloLens.Methods
Time-resolved 3D volumes and 2D multislice images were collected on a 3T MRI scanner (Skyra, Siemens Healthineers, Erlangen, Germany) using an undersampled radial FLASH acquisition. 3D volumes were collected in a short-axis orientation during diastole, and 2D multislice data consisted of 3 slices (two along the short axis, one in a 4-chamber orientation) collected without ECG gating. The following scan parameters were used for 3D imaging: 8 partitions/volume; 25% partition oversampling; partition thickness = 8mm; field-of-view = 300x300mm2; matrix = 128x128; in-plane resolution = 2.34x2.34mm2; flip angle = 7°; TR/TE = 2.92/0.73ms; bandwidth = 1115Hz/pixel; 30 coils. 2D imaging was performed with the same parameters except: slice thickness = 8mm; flip angle = 12°; TR/TE = 2.88/1.51ms; bandwidth = 1000Hz/pixel.
An acceleration factor of 9 (16/144 radial projections) was used, leading to an acquisition time of 467ms per 3D volume and 46ms per 2D slice (138ms for three slices). Fully-sampled calibration data (eight 3D measurements, 60 2D measurements) were collected during free-breathing without gating before accelerated imaging for the through-time radial GRAPPA reconstruction.
Images were reconstructed using a GPU-accelerated pipeline in the Gadgetron framework1,3. A coil compression step reduced the number of coils to 12. After reconstruction, images were exported over an Ethernet cable via a TCP/IP protocol to a computer running the HoloLens Unity software. 2D images were sent with position and orientation information such that slices were displayed in their relative positions, and the frame was updated once all three slices per frame were received. 3D volumes were rendered using a ray marching algorithm, and 2D slices were rendered using a texture shader. Images were cropped to a 90x90 matrix before display to better visualize the heart. The computer used to perform the GRAPPA reconstructions has an 8GB NVIDIA GeForce GTX 1080 graphics card; 10core, 2.2GHz Intel Xeon E5-2630 processor; and 64GB of 2400MHz DDR4 RAM. The computer running the Unity software has a 4GB NVIDIA Quadro K4200 graphics card; 14core, 2.6GHz Intel Xeon E5-2697v3 processor; and 64GB of 2400MHz RAM.
Results
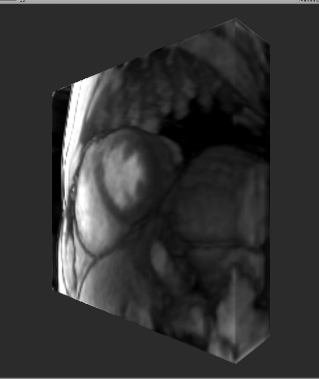
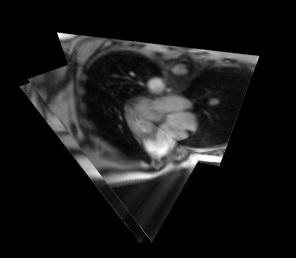
Examples of 3D volume and 2D multislice renderings are shown in Videos 1 and 2, respectively. Note that while these videos show the renderings rotating, a user wearing a HoloLens headset would control their view of the volume. The user is able to move around the hologram, and toward and away from it to view different structures from several angles. The time required for the steps in this system are given in Table 1. 3D volumes are acquired in 467ms/volume and displayed with a latency of 353ms. One frame of three 2D slices is acquired in 138ms/frame and displayed with a latency of 163ms.Discussion
This system provides rapid data acquisition, reconstruction, and intuitive visualization of a time series of 2D multislice or 3D MRI images as they change in real time. Highly undersampled data are acquired using a radial trajectory and reconstructed using a fast implementation of through-time radial GRAPPA1. Images are then displayed as holograms using the Microsoft HoloLens. Although the multislice dataset shown here has 3 slices, different numbers, positions, and orientations of slices can be selected to meet the needs of a particular procedure. Different orientations of 3D volumes can also be acquired. 2D imaging still has a total display latency that exceeds data acquisition, and future work will include reducing the reconstruction and rendering times for true real-time imaging. Future work also includes modifying the pipeline to handle 2D imaging interleaved with 3D imaging within the same session for increased user flexibility.Conclusion
By combining radial GRAPPA reconstruction, GPU acceleration, and HoloLens visualization, time-resolved 2D and 3D images can be acquired, reconstructed, and displayed in an intuitive holographic manner in real time.Acknowledgements
Siemens Healthcare, R01EB018108, NSF 1563805, R01DK098503, and R01HL094557.References
1. Franson D, Ahad J, Hamilton J, Lo W, Jiang Y, Chen Y, Seiberlich N. Real-time 3D cardiac MRI using through-time radial GRAPPA and GPU-enabled reconstruction pipelines in the Gadgetron framework. In: Proc. Intl. Soc. Mag. Reson. Med. 25.; 2017. p. 448.
2. Seiberlich N, Ehses P, Duerk J, Gilkeson R, Griswold M. Improved radial GRAPPA calibration for real-time free-breathing cardiac imaging. Magn. Reson. Med. 2011;65:492–505. doi: 10.1002/mrm.22618.
3. Hansen MS, Sørensen TS. Gadgetron: An open source framework for medical image reconstruction. Magn. Reson. Med. 2013;69:1768–1776. doi: 10.1002/mrm.24389.
Figures