0577
Application of a Scan-Specific Deep Learning Reconstruction to Multiband/SMS Imaging1Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States, 2Computer Assisted Clinical Medicine, University Medical Center Mannheim, Heidelberg University, Mannheim, Germany, 3Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN, United States
Synopsis
The use of convolutional neural networks (RAKI (Robust Artifical-neural-networks for k-space Interpolation)) trained for a scan specific acquisition is applied to Multiband/Simultaneous MultiSlice aliased k-space. With CNN’s of similar size as the RO-SENSE-GRAPPA kernels, reduced signal aliasing is obtained.
Introduction
Multiband/Simultaneous MultiSlice (MB/SMS) imaging has become an indispensable tool for accelerated acquisitions (1). Its combination with blipped CAIPI encoding (2) has enabled acquisitions with an undersampling of 6 or more. Such acceleration rates enables better coverage providing anatomically unique information (in diffusion) and improved temporal resolution of rapid brain dynamics (rs-fMRI). These acquisitions are typically reconstructed based on k-space interpolation. Such reconstruction approaches generate interpolation kernels from autocalibration signal (ACS), and the quality of ACS data is known to have a direct impact on the quality of the reconstructions (3). In improving coverage or resolution, even higher rates of acceleration are desirable. However, these are limited by noise amplification, as quantified by g-factors that are spatially varying and subject dependent, and the fidelity of the acquired data relative to the reference calibration.
In this work, we set to use a recently-proposed approach that tackles the k-space interpolation problem using nonlinear neural networks, called Robust Artifical-neural-networks for k-space Interpolation (RAKI) method (4) for MB/SMS imaging. RAKI replaces the linear convolutional kernels of MB/SMS algorithms with convolutional neural networks (CNNs) (5). It was shown to improve upon linear approaches in parallel imaging (6). Here, we modify RAKI for use with RO-SENSE-GRAPPA reconstruction (3,7), and evaluate its temporal stability in fMRI.
Methods
We employ a 3-layer CNN, which is trained in a scan-specific manner using the ACS data from the reference scan. As depicted in Figure 1, the three layers perform the following operations: F1(x) = ReLU(w1*x), F2(x) = ReLU(w2*x), F3(x) = w3*x, where * denotes convolution; w1,w2,w3 are convolutional kernels, and ReLU(x) = max(x, 0) is the rectified linear unit. Kernel sizes are chosen as: 4×3×2nc×128 for w1, 1×1×128×32 for w2, 2×3×32×(R–1) for w3, where R: MB/SMS factor, nc: number of coils, and factor of 2 is due to the real processing of complex data. Training is performed on ACS data using non-linear gradient descent with MSE as loss function. For comparison, RO-SENSE-GRAPPA (3,7) with 4×3 kernel size is implemented. Imaging was performed at 3T. The HCP protocol for functional imaging was used (1), with TE/TR=37/800ms, 2mm isotropic resolution, MB/SMS R=8, and blipped-CAIPI with FOV/3 shifts between adjacent multiband slices. Amount of ghosting and temporal stability were used to evaluate the reconstructions. Temporal stability was evaluated using TSNR, as the pixelwise mean relative to the standard deviation. For the linear reconstruction, signal leakage was further evaluated using the signal that is assigned to erroneous slices when the reconstruction is applied to a single band acquisition(8).Results
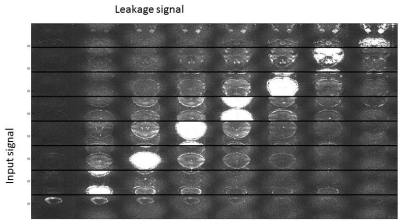
Figure 2 shows the reconstruction of a MB/SMS acquisition with RO-SENSE-GRAPPA (top) and RAKI (bottom). Residual ghosting is observed for most slices using the linear approach. For the RAKI reconstruction, these ghosts appear smaller. Figure 3 shows signal leakage from slice 6 into all other slices for RO-SENSE-GRAPPA, quantifying the trend observed with ghosting. Due to the non-linearity of RAKI, a similar analysis cannot be performed.
Figure 4 shows the TSNR maps for the RO-SENSE-GRAPPA reconstruction and the RAKI reconstruction, with similar performance for the two methods.
Discussion
The use of CNNs for multiband imaging has potential for enabling higher acceleration without increased signal leakage and with reduced noise amplification. The CNN-based approach suppresses common ghosting artifacts observed in the linear method.
The nonlinear processing of CNNs may also have the ability to perform a better reconstruction in the presence of data error effects or inaccurate signal behaviors such as pulsatile motion, which warrants further investigation.
In this study, we followed the original formulation of RAKI (4) in choosing small kernel sizes at each layer. The state-of-the-art works in MB/SMS use larger kernels, however this was not studied in this work.
Acknowledgements
Grant support: NIH P41 EB015894, NIH U01 EB025144, NIH R00 HL111410, and NSF CAREER CCF-1651825.References
1. Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E, Consortium WU-MH. The Human Connectome Project: a data acquisition perspective. Neuroimage 2012;62(4):2222-2231.
2. Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice Echo Planar Imaging with reduced g-factor penalty. Magn Reson Med 2012;67(5):1210-1224.
3. Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med 2010;63(5):1144-1153.
4. Mehmet Akçakaya SM, Sebastian Weingärtner, Kâmil Uğurbil. Scan-specific Robust Artifical-neural- networks for k-space Interpolation-based (RAKI) Reconstruction: Database-free Deep Learning for Fast Imaging. Magn Res Med 2018.
5. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436-444. 6. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47(6):1202-1210.
7. Koopmans PJ. Two-dimensional-NGC-SENSE-GRAPPA for fast, ghosting-robust reconstruction of in-plane and slice-accelerated blipped-CAIPI echo planar imaging. Magn Reson Med 2017;77(3):998-1009.
8. Cauley SF, Polimeni JR, Bhat H, Wald LL, Setsompop K. Interslice leakage artifact reduction technique for simultaneous multislice acquisitions. Magn Reson Med 2014;72(1):93-102.
Figures
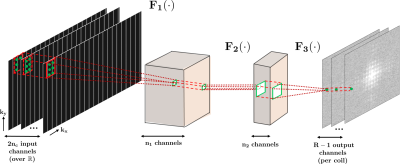
Figure 1: The three layer network structure used in this study for the CNN. The first layer takes in the sub-sampled zero-filled k-space (832×104×64), as embedded into the real field. The convolutional filters in this layer, w1 are of size 4×3×2nc×128. This is followed by a rectified linear unit (ReLU) operation. The second layer takes in the output of the first layer and applies convolution filters, denoted by w2, of size 1×1×128×32. This layer includes a ReLU operation. These two layers non-linearly combine the acquired k-space lines. The final layer produces the desired reconstruction output by applying convolutional filters, w3, of size 2×3×32×(R-1).
Figure 2: Reconstruction of 8 slices acquired simultaneously, with 1/3 FOV shift between adjacent slices. All images are windowed equally wrt. The maximum signal across all slices. Top row is with a 3x4 kernel (RO x PE), and shows residual ghosting outside. Bottom row shows that for a local kernel the RAKI shows more incoherent residual signal.
Figure 3. The TSNR (mean divided by standard deviation of the series) over 100 images for the 3 different methods.