0571
Inception-CS: Deep Learning For Sparse MR Reconstruction in Glioma Patients1University of California San Francisco, San Francisco, CA, United States, 2Columbia University Medical Center, New York, NY, United States, 3University of California Irvine, Irvine, CA, United States, 4Hofstra Northwell School of Medicine, Manhasset, NY, United States
Synopsis
Sparse MR image reconstruction through deep learning represents a promising novel solution with early results suggesting improved performance compared to standard techniques. However, given that neural networks reconstruct using a learned manifold of rich image priors, it is unclear how the algorithm will perform when exposed to pathology not present during network training. In this study we: (1) present a novel Inception-CS architecture for reconstruction using extensive residual Inception-v4 modules; (2) demonstrate state-of-the-art reconstruction performance in glioma patients however only when representative pathology is available during algorithm training.
Introduction
Convolutional neural networks (CNN) represent a new promising solution for sparse MR reconstruction. In just the past year, several studies have suggested that CNNs have the potential to outperform traditional methods based on speed and accuracy1–5. However these studies have investigated reconstruction in the absence of pathology, and it is unclear how the algorithm may perform when exposed to novel diseases not present during network training. This is important given that the advantages of CNN-based reconstruction are in part driven by a learned manifold of rich image priors.
In this study we investigate patients with glioma, the most common primary brain tumor6. Specifically, we compare reconstruction performance of two CNNs, one trained using only slices without tumor, and a second using the entire volume. To do this, we introduce a novel Inception-CS architecture optimized for compressed-sensing reconstruction by combining fully convolutional residual neural networks (FCR-NN) with extensive state-of-the-art residual Inception-v4 modules7,8.
Methods
Using open-source data available from The Cancer Imaging Archives9, glioma patients were identified and corresponding T1-weighted post-contrast as well as FLAIR acquisitions were downloaded. For each patient, a fully automated segmentation algorithm7 was used to identify slices containing tumor. All slices were zero-padded to a square matrix and resized to 128 x 128. Raw images were mapped to k-space using a Fourier transform (FFT) and subsampled with a variable density Poisson disc generated by the Berkeley Advanced Reconstruction Toolbox (BART) toolbox10.
A novel deep learning architecture named Inception-CS was designed for image reconstruction directly from subsampled k-space data. The key algorithm features arise from two observations: (1) given that reconstruction from k-space requires mapping dense input and output matrices, we exploit the efficiencies of a FCR-NN (U-net-based architecture11 with residual connections); (2) given that local frequency domain changes have global spatial domain effects, we incorporate extensive use of residual Inception-v4 modules8 to synthesize multi-resolution features at each intermediate block. Further details are provided in Figure 1.
Two neural networks were trained, one using only slices without glioma (non-tumor CNN), and a second containing the entire volume (combined CNN). Additionally, benchmark comparisons were made to wavelet sparsifying transform reconstructions with L1 regularization (λ = 0.01) at 500 iterations performed using the BART toolbox10. Reconstruction errors were assessed using a normalized mean square error (MSE) and compared on a slice-by-slice basis using a paired t-test.
Results
A total of 43,266 images from 268 glioma patients were identified, 7,342 of which contained tumor. A total of 36,102 images (228 patients) were used for training, and the remaining 7,164 images (40 patients) used for validation.
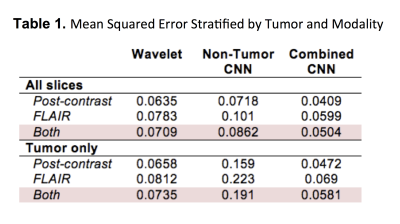
MSE for the non-tumor CNN was 0.0862 compared to an MSE of 0.0504 for the combined CNN (p<0.001), a difference that was exaggerated when comparing just tumor-containing slices (0.191 vs. 0.0581; p<0.001). By contrast, MSE for the wavelet transform was 0.0709, overall worse than the combined CNN (p<0.001) but significantly better than the non-tumor CNN (p=0.009). In general, FLAIR reconstruction resulted in higher MSE than post-contrast images for all models (p<0.001). Further stratification of results is shown in Table 1.
Difference maps highlighting reconstruction errors are shown in Figures 2-3. The average reconstruction time per slice for the non-tumor CNN, combined CNN and wavelet transform was 13.1 ms, 13.2 ms and 1690 ms respectively. Both CNN models were significantly faster than the wavelet transform (p<0.001).
Discussion
Our results are in line with previous studies that have suggested CNNs can be used for high performance sparse MR reconstruction1-5. Importantly however, we demonstrate that reliable CNN-based reconstruction of pathologic entities requires a representative distribution of training examples containing disease findings. Notably, a naïve CNN trained without pathology tends to favor “normal-appearing” reconstructions that obscure image abnormalities, with overall worse performance than a standard wavelet transform.
Differences in reconstruction errors are highlighted in Figures 2-3. Of note, reconstruction error is much more evenly distributed for the wavelet transform, whereas error tends to concentrate in specific image regions for CNNs. We hypothesize this in part relates to the relative ease in CNN-enabled modeling of common imaging features (e.g. large homogenous background regions and white matter patches) compared to more rare findings (e.g. localized pathology).
Conclusion
Sparse MR reconstruction using deep learning techniques has tremendous potential in outperforming conventional approaches, however state-of-the-art performance will likely require training on large datasets containing representative pathology. Given the expansive diversity of disease states, further work is necessary before application in routine clinical practice.Acknowledgements
The author of this paper gratefully acknowledges the support of NVIDIA Corporation with the donation of GPU hardware (GeForce GTX Titan X 12GB) used for this research.References
1. Wang, S. et al. Accelerating magnetic resonance imaging via deep learning. in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 514–517 (IEEE, 2016). doi:10.1109/ISBI.2016.7493320
2. Lee, D., Yoo, J. & Ye, J. C. Deep residual learning for compressed sensing MRI. in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 15–18 (IEEE, 2017). doi:10.1109/ISBI.2017.7950457
3. Mardani, M. et al. Deep Generative Adversarial Networks for Compressed Sensing (GANCS) Automates MRI. (2017).
4. Schlemper, J., Caballero, J., Hajnal, J. V, Price, A. & Rueckert, D. A Deep Cascade of Convolutional Neural Networks for MR Image Reconstruction. (2017). doi:10.1007/978-3-319-59050-9_51
5. Zhu, B., Liu, J. Z., Rosen, B. R. & Rosen, M. S. Image reconstruction by domain transform manifold learning. (2017).
6. Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016).
7. Chang, P. D. in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Second International Workshop, BrainLes 2016, with the Challenges on BRATS, ISLES and mTOP 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 17, 2016, Revised (eds. Crimi, A. et al.) 108–118 (Springer International Publishing, 2016). doi:10.1007/978-3-319-55524-9_11
8. Szegedy, C., Ioffe, S., Vanhoucke, V. & Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. (2016).
9. Clark, K. et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 26, 1045–1057 (2013).
10. Tamir, J. I., Ong, F., Cheng, J. Y., Uecker, M. & Lustig, M. Generalized Magnetic Resonance Image Reconstruction using The Berkeley Advanced Reconstruction Toolbox. (2016). doi:10.5281/zenodo.31907 11. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation.
Figures
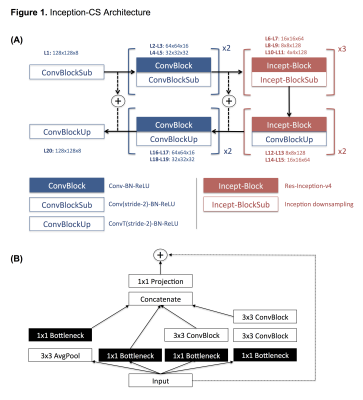
Figure 1. Inception-CS Architecture
(a) A U-net based fully convolutional residual neural network for efficient image reconstruction from raw subsampled k-space data. The network is composed of both collapsing and expanding arms combined by residual connections. (b) Extensive use of residual Inception-v4 modules is present throughout the deepest network layers for synthesis of multi-resolution features. Inception-v4 based architectures introduced by Google in the most recent ILSVRC challenges have yielded many of the current top-performing CNN algorithms; as far as the authors are aware, this is the first use of such design patterns for MR reconstruction.
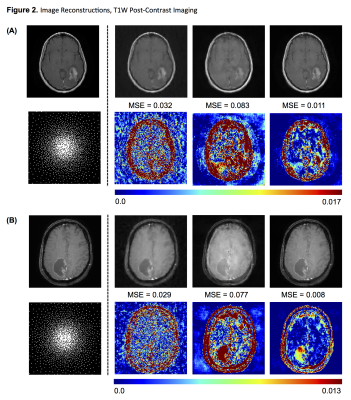
Figure 2. Image Reconstructions, T1W Post-Contrast Imaging
Two representative examples of reconstructions for T1-weighted post-contrast images containing enhancing glioma. From left to right: original image; sparsifying wavelet transform; CNN trained without tumor slices; CNN trained with entire dataset. Below each reconstruction is the corresponding difference map all shown with an identical scale to allow for comparison. In general, the combined CNN reconstructions outperform those based on wavelet transform, however the non-tumor CNN performs the worst.
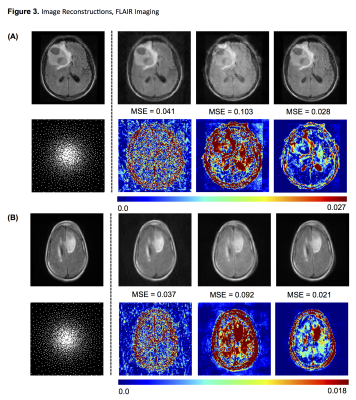
Figure 3. Image Reconstructions, FLAIR Imaging
Two representative examples of reconstructions for FLAIR images containing glioma. From left to right: original image; sparsifying wavelet transform; CNN trained without tumor slices; CNN trained with entire dataset. Below each reconstruction is the corresponding difference map all shown with an identical scale to allow for comparison. In general, the combined CNN reconstructions outperform those based on wavelet transform, however the non-tumor CNN performs the worst.