0475
Effects of reconstruction with deformable motion correction on abdominal DCE-MRI images1Radiation Oncology, University of Michigan, Ann Arbor, MI, United States, 2Radiology, University of Michigan, Ann Arbor, MI, United States, 3Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Synopsis
Image reconstruction with deformable motion correction for dynamic contrast-enhanced (DCE) MRI can counteract artifacts in dynamic images and estimated perfusion maps. In this study, we present the results of applying a reconstruction method with deformable motion correction to 54 DCE-MRI examinations of 31 patients. Deformable motion correction is found to compensate for up to 15 mm of residual motion after rigid-body motion correction. Motion-correction also reduces liver-wide bias caused by motion-distorted input functions and removes localized artifacts at liver edges. Finally, for several cases, motion-correction makes lesion edges sharper in reconstructed images.
Introduction
DCE MRI can be used to estimate liver perfusion and function
for patients with liver cancer (1,2). However, respiratory motion can
distort DCE-MRI uptake curves by introducing aliasing and blur into
reconstructed images. We have previously reported on a method to correct for respiratory
motion during reconstruction of abdominal DCE-MRI images (3). In this study, we present
the effects of this method on uptake curves and describe the type and range of
motion observed among patients.
Methods
Under IRB approval, 54 DCE-MRI examinations were performed with a 3T scanner on 31 patients (11 women, 20 men, age at examination 48–78) using a golden-angle stack-of-stars gradient-echo sequence as part of a pilot study of individualized adaptive radiotherapy for hepatocellular carcinoma. Following administration of 20 ml of Gd-BOPTA, patients were scanned for 5 minutes.
Raw data was collected after each examination and reconstructed using our motion-correction method (3). This method first reconstructs an image time series with high temporal but low spatial resolution and performs a rigid-body registration of the liver to produce a respiratory motion signal (4). Using this signal, radial spokes are sorted by breathing phase and reconstructed into 21 respiratory motion-state images. These are then aligned to the end-exhale state using deformable registration (5). The resulting deformation fields are used to deform single-spoke projection images to the exhale state before combining them using view sharing (4) into a dynamic DCE-MRI times series without respiratory motion but with the contrast-agent dynamics maintained. Images were also reconstructed by the same view-sharing technique but without deforming the projection images first.
For each view-sharing reconstruction, arterial and portal-venous input functions were extracted and arterial and portal-venous perfusion maps were estimated for the liver using a dual-input single-compartment model (6). In addition, deformation fields were compared to the best rigid-body registration transform for the liver.
Results
For the 54 examinations, the distances travelled by the center of the liver from end-exhale to end-inhale ranged from 8 mm to 46 mm in the SI direction, 2 mm to 10 mm in the LR direction and 3 mm to 35 mm in the AP direction. Median motion ranges were 15 mm, 4 mm and 8 mm for SI, LR, and AP respectively. After correction for rigid-body motion, the median residual displacement among liver voxels, as described by the deformation fields, ranged from 1 mm to 6 mm among examinations with a population mean of 2 mm. The 95th percentile of the residual displacement varied between 2 mm and 15 mm with a mean of 7 mm. The minimum Jacobian determinant inside the liver ranged from 0.73 to 0.94 and the maximum Jacobian determinant from 1.06 to 1.46.
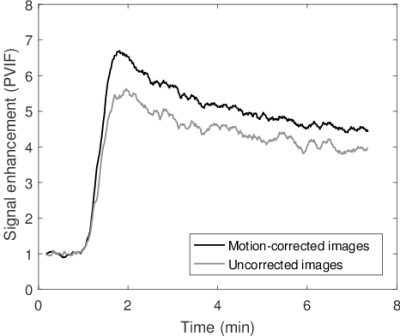
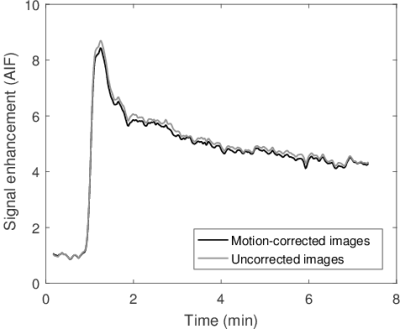
The peak amplitude of the portal-venous input function (PVIF) was significantly higher in motion corrected images compared to uncorrected (p << 0.01, mean increase: 9%). No significant difference was found for the peak amplitude of the arterial input function (AIF) (p = 0.16). Example PVIFs and AIFs for a subject are shown in Fig. 1 and Fig. 2.
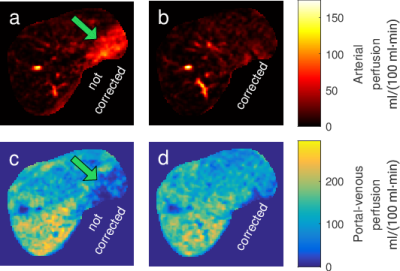
Median portal-venous perfusion in perfusion maps estimated from motion-corrected images was significantly lower (p < 0.01, CI = [1, 8.5] ml/(100 ml·min)) compared to perfusion maps from uncorrected images. No significant difference was found for median arterial perfusion between corrected and uncorrected images (p = 0.12). Some local artifacts in dynamic images were observed to propagate into local artifacts in perfusion maps as shown in Fig. 3.
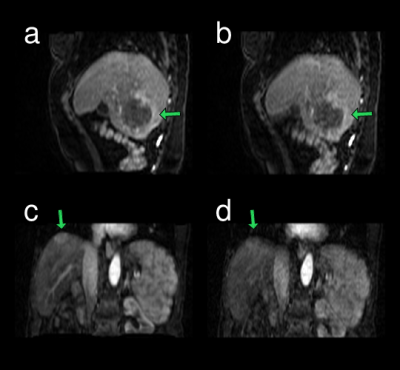
For several lesions, motion-correction made lesion borders sharper as illustrated in Fig. 4.
Discussion
Rigid-body motion correction cannot fully compensate for the movement of the liver and can leave mean residual displacements of up to 15 mm uncorrected for inside the liver. Non-rigid-body motion correction can therefore make it possible to extend motion compensation to all parts of the liver.
DCE-MRI reconstruction with integrated motion correction can restore uptake curves to higher peak amplitudes where motion is present. This is particularly apparent for the portal vein, which is a small structure with high contrast to surrounding liver tissue. The effect of reducing the peak amplitude of the portal vein is a systematic motion-dependent increase of the estimated portal-venous perfusion. Motion may also introduce local artifacts close to the liver edge that can be corrected for with the described method. For some lesions, motion correction improved the sharpness of the edge to surrounding tissue.
Conclusion
Motion-corrected reconstruction of DCE-MRI images can reduce
liver-wide bias in estimated perfusion maps caused by distorted input functions
as well as remove local artifacts and improve sharpness of lesions.Acknowledgements
This work was supported by NIH/NCI PO1 CA059827.References
1. Wang H, Farjam R, Feng M, Hussain H, Ten Haken RK, Lawrence TS, Cao Y. Arterial perfusion imaging-defined subvolume of intrahepatic cancer. Int J Radiat Oncol Biol Phys. 2014;89(1):167–74.
2. Cao Y, Wang H, Johnson TD, Pan C, Hussain H, Balter JM, Normolle D, Ben-Josef E, Ten Haken RK, Lawrence TS, Feng M. Prediction of liver function by using magnetic resonance-based portal venous perfusion imaging. Int J Radiat Oncol Biol Phys. 2013;85(1):258–63.
3. Johansson A, Balter J, Cao Y. Motion-corrected image reconstruction of abdominal DCE-MRI images. Proc Intl Soc Mag Reson Med. 2016;25:1295.
4. Johansson A, Balter J, Cao Y. Rigid-body motion correction of the liver in image reconstruction for golden-angle stack-of-stars DCE MRI. Magn Reson Med. 2017;0(May):1–9 [Epub ahead of print].
5. Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Comput Meth Prog Bio. 2010;98(3):278–84.
6. Wang H, Cao Y. GPU-accelerated voxelwise hepatic perfusion quantification. Phys Med Biol. 2012;57(17):5601–16.
Figures