0473
Assessment of respiratory motion-resolved and nonrigid motion-corrected 3D Cartesian coronary MRA1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom
Synopsis
Slow acquisitions and susceptibility to respiratory motion artifacts are major challenges in free-breathing 3D whole-heart coronary MR angiography (CMRA). Recently, a respiratory-resolved approach has been proposed to improve scan efficiency and reduce motion artifacts using non-Cartesian acquisitions. However, irregular respirations compromise its suitability for Cartesian imaging. Here, sparsity in a motion-corrected domain is exploited to generate high-quality respiratory-resolved Cartesian images, used to estimate nonrigid motion fields. These are incorporated into a motion-corrected generalized matrix reconstruction, to further improve coronary vessel sharpness. Thus, this approach provides high-quality respiratory-resolved Cartesian CMRA images and a motion-corrected CMRA image at a given respiratory phase.
Introduction
Respiratory motion remains a major challenge in free-breathing 3D whole-heart coronary MR angiography (CMRA). Diaphragmatic navigator gating and tracking is commonly used to reduce motion artefacts. However, this approach only compensates for superior-inferior (SI) translational motion and has low scan efficiency, leading to prolonged scan times. Recently, XD-GRASP1,2 was proposed to achieve 100% scan efficiency. This method distributes the acquired data into multiple respiratory phases (or bins) and exploits Total Variation (TV) sparsity in the respiratory dimension to generate respiratory-resolved images. However, this technique may not be directly suitable for Cartesian imaging, particularly in the presence of irregular breathing patterns and intra-bin residual motion.3 Here, an optimized respiratory motion-resolved and motion-corrected two-step approach is proposed for Cartesian CMRA. The proposed Optimized Respiratory-resolved Cartesian Coronary MR Angiography (XD-ORCCA) incorporates motion information to increase the sparsity in the respiratory dimension.4 Additionally, 2D beat-to-beat translational motion estimated from image navigators5 (iNAVs) is used to minimize intra-bin motion. A second motion-corrected dataset is obtained estimating 3D bin-to-bin nonrigid motion from the XD-ORCCA images, which is incorporated into the general matrix description reconstruction.6,7 The proposed nonrigid motion-corrected reconstruction (MC-ORCCA) was compared against the highest quality bin of motion-resolved XD-GRASP reconstructions in six healthy subjects and two patients with cardiovascular disease.Methods
3D CMRA data is acquired using a prototype golden-step spiral-like Cartesian trajectory,8 which samples the ky-kz plane with spiral interleaves on a Cartesian grid. Consecutive spirals are separated by the golden-angle. Low-resolution 2D iNAVs are acquired at every heartbeat, before each spiral interleaf of the 3D CMRA acquisition. The 2D iNAVS are used to estimate beat-to beat 2D translational (SI and right-left: RL) motion and obtain the respiratory signal, which is used to distribute the 3D CMRA data into five equally populated respiratory bins. Each undersampled bin is reconstructed using 1) XD-GRASP and 2) XD-ORCCA. The latter is followed by a 3) nonrigid motion-compensated MC-ORCCA reconstruction.
The respiratory-resolved images $$$\hat{\rm{\mathbf{x}}}_{\rm{b}}$$$ were obtained by solving: 1) $$$\hat{\rm{\mathbf{x}}}_{\rm{b}}=\rm\arg\min\limits_{\rm{\mathbf{x}}_b}\left\{ \frac{1}{2}\left\|\mathbf {E}\,\mathbf{x}_b-\mathbf{k}_b\right\|_2^2 +\alpha\,\Psi_t(\mathbf{x}_b)\right\}$$$ and 2) $$$\hat{\rm{\mathbf{x}}}_{\rm{b}}=\rm\arg\min\limits_{\rm{\mathbf{x}}_b}\left\{ \frac{1}{2}\left\|\mathbf {E}\,\mathbf{x}_b-\mathbf{d}_b\right\|_2^2 +\alpha\,\Psi_t(\mathbf{x}_b)+\beta\,\Psi_s(\mathbf{x}_b)\right\}$$$, where $$$\mathbf{k}_b$$$ is the binned k-space data, $$$\mathbf{d}_b$$$ is the 2D translational corrected binned k-space data, $$$\Psi_{\rm{s}}$$$ is the 3D spatial TV function, $$$\alpha$$$ and $$$\beta$$$ are regularization parameters, $$$\mathcal{R}\rm{\mathbf{x}}_b=\mathit{T}_{b}\mathbf{x}_b$$$ is the motion-corrected domain, where $$$\mathit{T}_{\rm{b}}$$$ is the translation transform that maps the bin image $$$\rm{\mathbf{x}}_b$$$ to the reference image $$$\rm{\mathbf{x}}_1$$$ (end-expiration), and $$$\Psi_{\rm{t}}=\rm{\mathbf{x}}_1-\mathit{T}_{b}\mathbf{x}_b$$$ is the 1D temporal TV function. The operator $$$\mathbf{E}=\mathbf{A}_{\rm{b}}\mathbf{FS}$$$ incorporates the sampling matrix $$$\bf{A}$$$ for each bin b, Fourier transform $$$\bf{F}$$$ and coils sensitivities $$$\bf{S}$$$. The MC-ORCCA reconstruction was obtained by solving 3) $$$\hat{\rm{\mathbf{x}}}=\rm\arg\min\limits_{\rm{\mathbf{x}}}\left\{ \frac{1}{2}\left\|\mathbf {G}\,\mathbf{x}-\mathbf{d}\right\|_2^2\right\}$$$, where $$$\hat{\rm{\mathbf{x}}}$$$ is the motion corrected image, $$$\mathbf{G}=\sum\limits_{\rm{b}}\mathbf{A}_{\rm{b}}\mathbf{FS}\mathbf{U}_{\rm{b}}$$$ and $$$\mathbf{U}_{\rm{b}}$$$ are the nonrigid motion fields estimated (via image registration) from XD-ORCCA reconstructed $$$\hat{\rm{\mathbf{x}}}_{\rm{b}}$$$ images. These problems were solved with a (non)-linear conjugate gradient method.
In-vivo free-breathing experiments were performed on six healthy subjects and two patients on a 1.5T scanner (Siemens Magnetom Aera) with 18-channel body and 32-channel spine coils. 3D CMRA bSSFP acquisitions were performed using the following parameters: coronal orientation, FOV=320x320x80-104mm3, resolution=1x1x2mm3 (1.2x1.2x1.2mm3, for patients), TR/TE=3.6/1.56ms, flip angle=90°, T2 preparation (40ms), SPIR-like fat saturation, subject specific mid-diastolic trigger delay, acquisition window ~100ms, 1 spiral interleaf per heartbeat, with acquisition time of 9-12min. For the 2D iNAV acquisition, 14 bSSFP startup echoes were used (same geometry). Patient acquisitions were performed after gadolinium-based contrast injection. Patient 1 had a non-ischemic cardiomyopathy and Patient 2 had known ischemic heart disease with previous stent deployment in the mid-distal segment of the right coronary artery.
Results
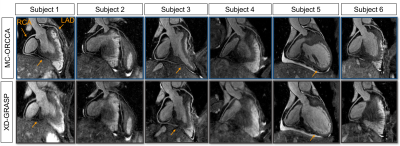
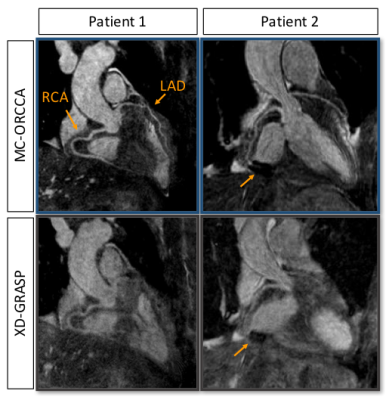
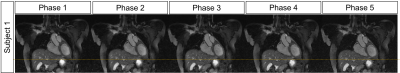
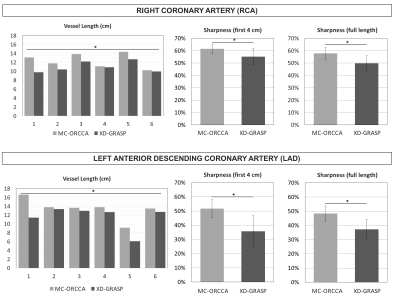
Reformatted images obtained with the proposed MC-ORCCA and XD-GRASP (respiratory phase with highest quality) are displayed in Fig.1 for all six healthy subjects. Motion blurring is observed in XD-GRASP Cartesian images for subjects with more irregular breathing patterns. This is particularly evident for subject 1 and patients (Fig.2). Overall, MC-ORCCA shows improvements in sharpness and visualization of distal coronary sections. The motion fields estimated from XD-ORCCA can be re-used to provide respiratory-resolved 3D motion-compensated images (Fig.3). Significant differences in the vessel length and sharpness were identified for both coronary arteries between MC-ORCCA and XD-GRASP approaches (Fig.4).Conclusions
The proposed nonrigid motion-corrected MC-ORCCA reconstruction framework provides high-quality 3D whole-heart images (single or multiple respiratory phases), from highly-efficient Cartesian acquisitions. 2D beat-to-beat translational motion is estimated from iNAVs and incorporated into the XD-ORCCA reconstruction to increase the sparsity in the respiratory dimension. Additionally, this motion information is used to compensate for residual intra-bin motion. XD-ORCCA provides high-quality respiratory-resolved images, essential for reliable nonrigid motion estimation, and thus, motion-corrected MC-ORCCA reconstruction.Acknowledgements
This work was supported by the following grants: EPSRC EP/N009258/1, EP/P032311/1 and EP/P001009/1, EP/P007619/1 and FONDECYT N° 1161051.References
1. Feng L, Axel L, Chandarana H, et al. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimension using compressed sensing. MRM 2016; 75:775-788.
2. Piccini D, Feng L, Bonanno G, et al. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. MRM 2017; 77:1473-1484.
3. Feng L, Chandarana H, Zhao T, et al. Golden-angle sparse liver imaging: radial or Cartesian sampling?. ISMRM 2017; #1285.
4. Correia T, Ginami G, Neji R, et al. Optimized respiratory-resolved motion-compensated 3D Cartesian coronary MRA. ISMRM 2018 (submitted).
5. Henningsson M, Koken P, Stehning S, et al. Whole-heart coronary MR angiography with 2D self-navigated image reconstruction. MRM 2012; 67:437-445.
6. Batchelor P, Atkinson D, Irarrazaval P, et al. Matrix description of general motion correction applied to multishot images. MRM 2005; 54:1273-1280.
7. Cruz G, Atkinson D, Henningsson M, et al. Highly efficient nonrigid motion-corrected 3D whole-heart coronary vessel wall imaging. MRM 2017; 77:1894-1908.
8. Prieto C, Doneva M, Usman M, et al. Highly efficient respiratory motion compensated free-breathing coronary MRA using golden-step Cartesian acquisition. JMRI 2015; 41:738-746.
Figures