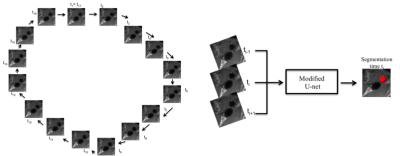0439
Towards a fully Automated Time-context Sensitive Convolutional Neural Network for Common Carotid Artery Lumen Segmentation on Dynamic MRI1Seaman Family Magnetic Resonance Research Centre, Calgary, AB, Canada, 2Medical Image Computing Lab, Campinas, Brazil
Synopsis
Carotid artery atherosclerosis is one of the main causes of stroke and there is a pressing need for a non-invasive method to quantify, monitor and assess carotid artery stenosis, composition and distensiblity. Here we focus on developing a fully automated convolutional neural network (CNN) with time-context for segmenting the common carotid artery lumen from dynamic magnetic resonance images. The challenge in developing a fully automated carotid segmentation algorithm is that there are other vessels with size and spatial location comparable to the carotid artery. Our preliminary results indicate that a CNN with time-context is capable of distinguishing and segmenting the carotid artery from other vessels.
Introduction
Carotid artery atherosclerosis is one of the main causes of stroke. Although the current standard guideline for treatment is just stenosis percentage1, the arterial health is characterized by a more complex mechanism that depends on carotid artery stenosis, wall composition and vessel distensibility. Cine fast spin echo (FSE)2 is a cardiac-gated magnetic resonance (MR) imaging method that acquires dynamic images throughout the cardiac cycle with little additional MR scan time. The k-space data acquired during the cine FSE scan is used to reconstruct static high-resolution images, and by drawing on 4D (3D space + time) sparsity and compressed sensing algorithms, allows the reconstruction of high-resolution cardiac cycle-resolved images. This work presents preliminary results towards developing a fully automated common carotid artery (CCA) lumen segmentation tool based on a convolutional neural network (CNN) with time-context from the cardiac cycle. This tool will allow artery distensibility characterization, and is a component of a larger project with the overall goal to understand the impact of carotid stenosis, composition and distensibility on normal aging and cerebrovascular disease.Materials and Methods
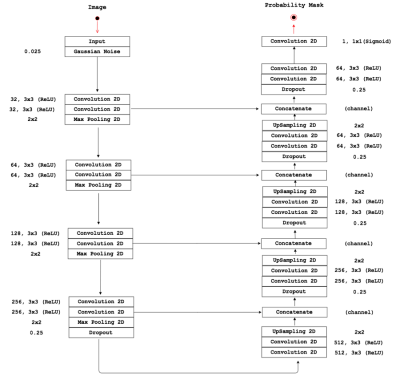
The carotid arteries in twelve healthy subjects were scanned using cine FSE. Ten subjects were scanned five times and two subjects were scanned only once. The images were acquired on a 3 T scanner (Discovery 750; General Electric, Waukesha, WI) and reconstructed at 16 phases across the cardiac cycle. The images were manually annotated by using an initial segmentation obtained using a morphology-based semi-automatic tool3. Our CNN approach used a 2D U-net4 architecture with dropout layers and Gaussian noise in the input (Figure 1). The time-context was obtained by encoding temporally adjacent images into three channels (used for RGB encoding). The first channel contains the image at time ti-1, the second channel at ti (i.e., the time being segmented), and the last channel at ti+1 (Figure 2). Due to the cyclic nature of the cardiac cycle, the last time point wrap around to the first (Figure 2). We used a leave one subject out cross-validation to test our approach. The CNN was trained using 64x64 patches combined with data augmentation techniques5. For each image slice, twenty patches were extracted that included the carotid artery (positive samples) and fifteen patches were extracted from surrounding regions without the carotid artery (negative samples). These parameters were determined experimentally by testing different patch sizes and distributions of negative and positive samples. The source code and details of the training are available (https://github.com/rmsouza01/carotid-cnn). The automated results were compared against the manual segmentations using sensitivity, specificity and the Dice coefficient (a measure of overlap)6 as metrics; all range between 0 and 1.Results and Discussion
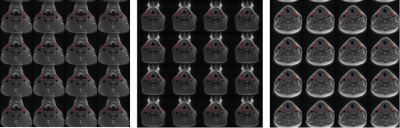
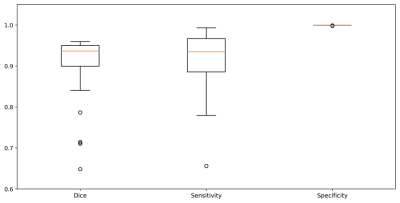
Our results can be divided in three performance groups (Figure 3): 1) only the carotid arteries were segmented (15 cases, 29%); 2) the carotid arteries and other vessels (of similar size, shape and location as the carotid arteries) were segmented (15 cases, 29%)); 3) the carotid arteries were correctly segmented along with other smaller structures that could be removed by post-processing using an area-open filter7 (22 cases, 42%). In all 52 cases, the carotid arteries were segmented. These results are a first step towards a fully automated CCA lumen segmentation tool. Although, we used 52 image sets, they come from only 12 different subjects, which limited the amount of anatomic variability that the CNN has available during its learning stage. The sensitivity, specificity and Dice coefficient results of our method, considering only the structures corresponding to the carotid artery, are summarized in Figure 4. The state-of-the-art Dice coefficient for CCA lumen segmentation3,8 is about 0.93 using semi-automatic methods that do not use time-context information. Our approach achieved a Dice coefficient of 0.91. Unlike the other techniques, however, our method aims towards being fully automated and profits from the time-context information. We expect that with a larger dataset our method will achieve comparable results with the state-of-the-art with the added advantage of being fully automated.Conclusions
We presented a CNN that capitalizes on time-context information to perform automatic segmentation of the CCA lumen in dynamic MR images. Using a small dataset with 52 MR acquisitions (12 healthy subjects), we were able to achieve a result comparable to the state-of-the-art (Dice coefficient: 0.91 vs 0.93). Our method successfully segmented 37 of the 52 (71%) images without the necessity of user interaction. As future work, we want to investigate this architecture using a larger dataset composed of both healthy and atherosclerotic subjects. This tool will be used inside a larger study to characterize carotid artery distensibility, plaque composition and stenosis to try to understand normal aging and occurrence of cerebrovascular disease.Acknowledgements
The authors would like to thank Hotchkiss Brain Institute; CAPES process PVE 88881.062158/2014-01; FAPESP CEPID2013/07559-3 and the CREATE I3T program for providing financial support.References
1. Rothwell P, Eliasziw M, Gutnikov S, et al. Analysis of pooled data from the randomised controlled trials of endarterectomy for symptomatic carotid stenosis. The Lancet 2003;361(9352):107-16
2. Boesen ME, Maior Neto LAS, Pulwicki A, Yerly J, Lebel RM, Frayne R. Fast spin echo imaging of carotid artery dynamics: FSE of Carotid Artery Dynamics. Magnetic Resonance in Medicine 2015;74(4):1103-09
3. Rodrigues L, Souza R, Rittner L, Frayne R, Lotufo R. Common Carotid Artery Lumen Segmentation from Cardiac Cycle-resolved Cine Fast Spin Echo Magnetic Resonance Imaging. Conference on Graphics, Patterns and Images (SIBGRAPI). Niterói, Rio de Janeiro, Brazil, 2017.
4. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2015. Springer.
5. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems; 2012.
6. Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol. Skr. 1948;5:1-34
7. Vincent L. Morphological area openings and closings for grey-scale images. Shape in Picture: Springer, 1994:197-208.
8. Ukwatta E, Yuan J, Rajchl M, Qiu W, Tessier D, Fenster A. 3-D carotid multi-region MRI segmentation by globally optimal evolution of coupled surfaces. IEEE transactions on medical imaging 2013;32(4):770-85
Figures