0433
Deep learning diffusion fingerprinting to detect brain tumour response to chemotherapy1Centre for Advanced Biomedical Imaging, University College London, London, United Kingdom
Synopsis
Convolutional neural networks (CNNs) often require very large datasets for robust training and evaluation. As an alternative approach, we introduce deep learning diffusion fingerprinting (DLDF), which treats every voxel as an independent data point, rather than using whole images or patches. We use DLDF to classify diffusion-weighted imaging voxels in a mouse model of glioblastoma, both prior to and in response to Temozolomide chemotherapy. We show that, even with limited training, DLDF can automatically segment brain tumours from normal brain, can distinguish between young and older tumours and that DLDF can detect if a tumour has been treated with chemotherapy.
Introduction
Convolutional neural networks (CNNs) are by far the most popular type of deep learning architecture and they are widely used for a range of biomedical imaging applications [1]. CNNs allow subtle features to be characterised, but often require very large, labelled data sets, and they require entire images (or patches from images) as the input into the neural network. In multi-direction, multi-b-value diffusion MRI acquisitions, the signal (or ‘diffusion fingerprint’) from within a single voxel contains subtle variations caused by the underlying tissue microstructure. We propose deep learning diffusion fingerprinting (DLDF), which uses the diffusion fingerprint from each voxel as an independent data point to train a deep neural network. This approach aims to produce diagnostic information with a relatively small number of subjects. In this abstract, we investigate the ability of DLDF to distinguish brain, background, control and treated tumour pixels, independent of pixel location.Methods
Glioma mouse model and therapy: 24 mice were injected with GL261 glioma cells in the right striatum. After 13 days of growth, baseline scans were acquired in all mice (day 0). Temozolomide (TMZ) was administered to the treated group of mice (n = 12). Vehicle was administered to the control group (n = 12). After nine more days (day 9), follow-up images were acquired.
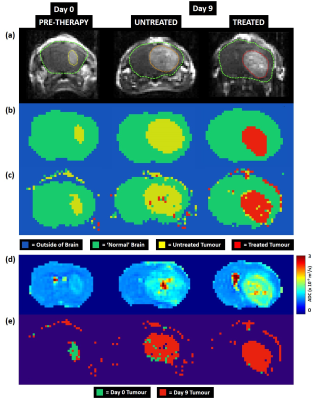
MRI: A PGSE diffusion-rich dataset with 46 diffusion weightings and a 42-direction DTI was acquired on a 9.4T Agilent scanner [2]. Five coronal slices were acquired in each animal. Slice-matched, T1-weighted post-Gd-DTPA images were acquired (Figure 1a) after diffusion MRI.
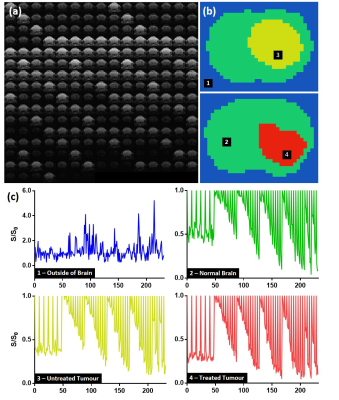
Deep Learning: For training the neural network, four categories were manually identified on Gd-DTPA images: 1) outside of the brain, 2) normal-appearing brain, 3) untreated tumour at day 0 and day 9, 4) treated tumour at day 9 (Figure 1b). Diffusion fingerprints were produced for each pixel consisting of 232 ordered data points (Figure 2). Python libraries (Keras and TensorFlow [3]) were used to construct a neural network with 5 hidden layers. Each hidden layer was regularized with 20% dropout, and used ReLU activation and normally-distributed initialisation. 50% of the data (6 control, 6 treated) were randomly assigned to a training set, which were further refined to include equal numbers of fingerprints from each category (total number of fingerprints = 6,608). Neural network training was performed using stochastic gradient-descent optimisation with a mean squared error loss function. Training was performed for 50,000 epochs, with 10,000 steps in each. 5 animals per group were used for evaluation.
Results
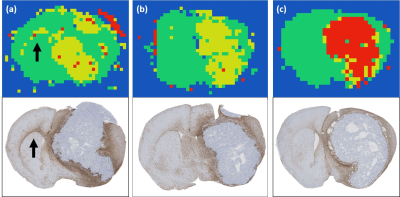
For training, the T1-weighted Gd-DTPA images (Figure 1a) were used to create category masks (Figure 1b) which were applied to the multi-dimensional DWI images (Figure 2a) to generate pixel-wise diffusion fingerprints (Figure 2c). Upon visual inspection of the diffusion fingerprints, some subtle differences could be observed between fingerprints from normal brain (green) and tumour (yellow/red).
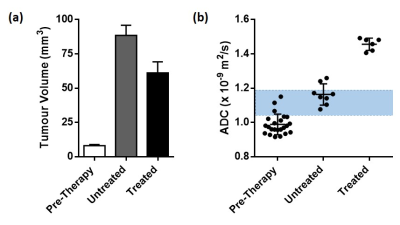
The neural network training accuracy was 98.65%. The evaluation accuracy was 98.54%. A strong agreement was observed between manually segmented category masks (Figure 1b) and the evaluation DLDF maps (Figure 1c), demonstrating the ability of the neural network to automatically segment the tumours. The neural network was also able to distinguish between tumours based on age (Figure 1e) – voxels within day 0/day 9 tumours were predominantly classified as day 0/day 9 tumours, respectively. In comparison, ADC alone was unable to stratify all pre-therapy tumours from day 9 tumours (Figure 3).
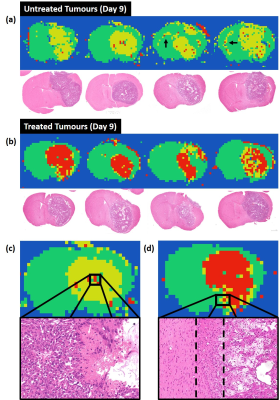
DLDF category maps at day 9 showed a strong spatial accordance with histological slices (Figure 4). For example, treated tumours generally showed a distinctive rim of ‘untreated’ pixels (yellow) at the periphery of the tumour, which could correspond to regions in H&E images where tumour transitions into the normal brain (Figure 4d). In some untreated brains, the DLDF category maps showed tumour invasion into the contralateral hemisphere along the corpus callosum, which also showed enhanced GFAP-staining on histology (Figure 5).
Discussion
We have shown that DLDF can automatically segment tumours from normal brain, DLDF can automatically distinguish between young and older (after 9 days of growth) tumours and that DLDF can detect whether or not a tumour has been treated with chemotherapy. We have also shown that, spatially, the category maps generated by DLDF were broadly consistent with histology and that interesting features are detected – these include regions which are possibly less sensitive to Temozolomide treatment and transitional regions where the microstructure may be a mixture of normal brain and tumour cells. Our results show the potential for applying deep learning on an individual voxel-wise level, and could be applied to many other types of multi-dimensional acquisitions.Acknowledgements
No acknowledgement found.References
[1] Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. arXiv preprint arXiv:170205747 (2017)
[2] Panagiotaki E, Walker-Samuel S, Siow B, et al. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Research (2014).
[3] Abadi M, Agarwal A, Barham P, et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:160304467 (2016)
Figures