0429
Gibbs-Ringing Artifact Reduction in MRI via Machine Learning Using Convolutional Neural Network1Guangdong Provincial Key Laboratory of Medical Image Processing, School of Biomedical Engineering, Southern Medical University, Guangzhou, China, 2Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 3Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China
Synopsis
The Gibbs-ringing artifact is caused by the insufficient sampling of the high frequency data. Existing methods generally exploit smooth constraints to reduce intensity oscillations near high-contrast boundaries but at the cost of blurring details. This work presents a convolutional neural network (CNN) method that maps ringing images to their ringing-free counterparts for Gibbs-ringing artifact removal in MRI. The experimental results demonstrate that the proposed method can effectively remove Gibbs-ringing without introducing noticeable blurring.
Purpose
To develop a machine learning based approach to remove Gibbs-ringing artifact in MRI.Introduction
Gibbs-ringing artifact refers to a series of spurious intensity oscillations near sharp edges in MR images.1 This artifact is caused by the insufficient sampling of high-frequency data in the k-space domain. In practice, the ringing artifact degrades image quality, and can complicate diagnosis and affect the subsequent parameter quantification.2,3 To tackle this problem, model-based data extrapolation or image smoothing approaches have been developed, and they can reduce this artifact but at the expense of blurring.4,5
Machine learning methods such as autoregressive modeling and multilayer neural network have been trained on known truncated data set and then used to predict unknown high frequency components from the measured low frequency data.6-8 Motivated by the CNN for compression artifact reduction,9 this work aims to develop a CNN-based method for Gibbs-ringing artifact reduction in MRI.
Methods
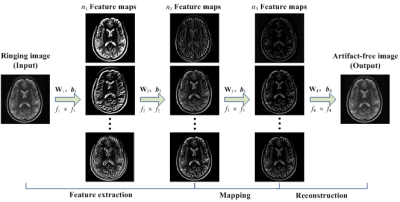
Our proposed method employs a CNN that directly learns an end-to-end mapping from a ringing image to a ringing-free image, and then obtain the final output by replacing the low-frequency part of the CNN-estimated image with the measured k-spaced data. We name the proposed method as Gibbs-ringing artifact reduction using CNN (GRA-CNN). The overall architecture of GRA-CNN is shown in Figure 1. GRA-CNN consisted of a four-layer convolutional network. Rectified linear units (ReLU, max(0,x))10 was applied as the activation function, and batch normalization11 was used to accelerate network training and boost accuracy. The filter size was set f1 = 9, f2 = 7, f3 = 1, and f4 = 5; and the filter number was set n1 = 64, n2 = 32, n3 = 16 and n4 = 1. The network parameters Θ = {Wi, bi, i = 1,2,3,4} were learned for the end-to-end mapping function. The mean-squared error (MSE) was used as the loss function and minimized by using stochastic gradient descent with the standard backpropagation.
The training data consisted of 17532 T2-weighted (T2W) brain images of 136 healthy adult subjects from the human connectome project (http://www.humanconnectome.org, 900 Subjects Data). Testing data consisted of T2W and diffusion-weighted (DW) images. The T2W images of normal brain were acquired on a 1.5T MR scanner (OPTIMA MR360, GE, America) using the fast-spin-echo sequence: TR/TE=5900/117 ms, FOV=24×24 cm2, pixel size=0.47×0.47 mm2, and slice thickness=6.0 mm. The brain tumor were acquired on a 3T scanner (Achieva, Philips, Netherlands) using the fast-spin-echo sequence: TR/TE = 3000/80 ms, FOV=22×22 cm2, pixel size=0.43×0.43 mm2, and slice thickness=6.0 mm. The DW data was acquired on a 3T scanner (Achieva, Philips, Netherlands) using an interleaved EPI sequence: TR/TE=3000/83 ms, FOV=22×22 cm2, pixel size=1.0×1.0 mm2, slice thickness=6.0 mm, shots=6, and b-value=800 s/mm2, and diffusion directions=10.
We implemented the GRA-CNN network using the MatConvNet package [46]. In the training, the convolution parameters were randomly initialized from a Gaussian distribution with a standard deviation of 0.001, and the learning rate was set 10-4. The root-mean-square error (RMSE), peak signal-to-noise ratio (PSNR) and structure similarity (SSIM) index were calculated for the quantitative evaluation.
Results
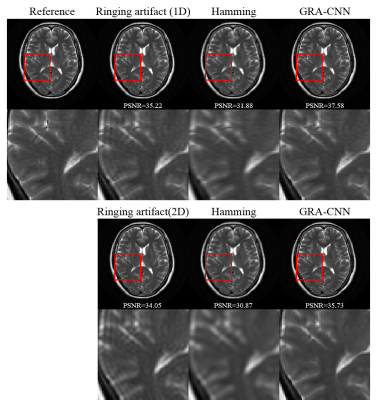
Figure 2 presents the results of Gibbs-ringing reduction on a representative T2W normal brain images with 50% data truncation in one dimension and both dimensions. The low-pass hamming filter removes the ringing artifact, but shows a smoothing of rapidly changing details and reduces PSNR. In contrast, GRA-CNN effectively eliminates the ringing artifact without noticeable blurring.
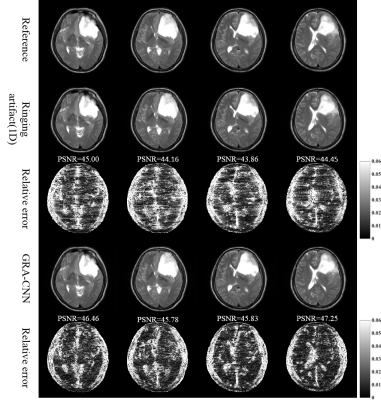
The application of the proposed method on the T2W images of brain tumor is presented in Figure 3. The GRA-CNN model trained by using normal brain images also successfully removes ringing artifact in brain tumors, without noticeable blurring.
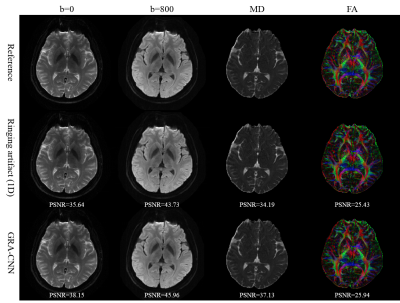
Figure 4 shows the application of the proposed method to DW images. The ringing artifacts are successfully removed from DW images. The mean diffusivity (MD) map is significantly improved by the proposed method, and the fractional anisotropy (FA) maps before and after artifact suppression are comparable.
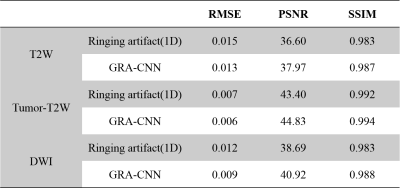
Table 1 presents the quantitative evaluation of the proposed method. The GRA-CNN method consistently generates images with improved RMSE, PSNR and SSIM values, for both T2- and diffusion-weighted images.
Discussion & Conclusion
This work presents a novel and simple method that employs a four-layer CNN as an end-to-end mapping that inputs ringing MR image and outputs the artifact-free one to remove Gibbs-ring artifact from MR images. The results demonstrate that the proposed GRA-CNN method can effectively remove Gibbs-ringing artifacts without noticeable blurring.Acknowledgements
No acknowledgement found.References
- Wood, M.L. and R.M. Henkelman, Truncation artifacts in magnetic resonance imaging. Magn Reson Med, 1985. 2(6): p. 517-26.
- Ferreira, P., et al., Variability of myocardial perfusion dark rim Gibbs artifacts due to sub-pixel shifts. J Cardiovasc Magn Reson, 2009. 11: p. 17.
- Veraart, J., et al., Gibbs ringing in diffusion MRI. Magn Reson Med, 2015.
- Amartur, S. and E.M. Haacke, Modified iterative model based on data extrapolation method to reduce Gibbs ringing. J Magn Reson Imaging, 1991. 1(3): p. 307-17.
- Constable, R.T. and R.M. Henkelman, Data extrapolation for truncation artifact removal. Magn Reson Med, 1991. 17(1): p. 108-18.
- Smith, M.R., et al., Application of autoregressive modelling in magnetic resonance imaging to remove noise and truncation artifacts. Magn Reson Imaging, 1986. 4(3): p. 257-61.
- Barone, P. and G. Sebastiani, A new method of magnetic resonance image reconstruction with short acquisition time and truncation artifact reduction. IEEE Trans Med Imaging, 1992. 11(2): p. 250-9.
- Yan, H. and J. Mao, Data truncation artifact reduction in MR imaging using a multilayer neural network. IEEE Trans Med Imaging, 1993. 12(1): p. 73-7.
- Dong, C., et al. Compression artifacts reduction by a deep convolutional network. in Proceedings of the IEEE International Conference on Computer Vision. 2015.
- Nair, V. and G.E. Hinton. Rectified linear units improve restricted boltzmann machines. in Proceedings of the 27th international conference on machine learning (ICML-10). 2010.
- Ioffe, S. and C. Szegedy. Batch normalization: Accelerating deep network training by reducing internal covariate shift. in International Conference on Machine Learning. 2015.
Figures