0428
Deep Learning Method for Non-Cartesian Off-resonance Artifact Correction1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Synopsis
3D cones trajectories have the flexibility to be more scan-time efficient than 3D Cartesian trajectories, especially with long readouts. However, long readouts are subject to blurring from off-resonance, limiting the efficiency. We propose a convolutional residual network to correct for off-resonance artifacts to allow for reduced scan time. Fifteen exams were acquired with both conservative readout durations and readouts 2.4x as long. Long-readout images were corrected with the proposed method. The corrected long-readout images had non-inferior (p<0.01) reader scores in all features examined compared to conservative readout images.
Introduction
3D cones trajectories have the flexibility to be more scan-time efficient compared to 3D Cartesian trajectories but respond to off-resonance with blurring rather than shifts1. The blurring is especially apparent when long readouts are used to reduce scan time. Existing methods for addressing off-resonance are too computationally slow for clinical viability. Therefore, we investigated a fast deep learning method for correcting off-resonance artifacts to enable longer readouts for reducing scan time.Methods
Dataset Creation
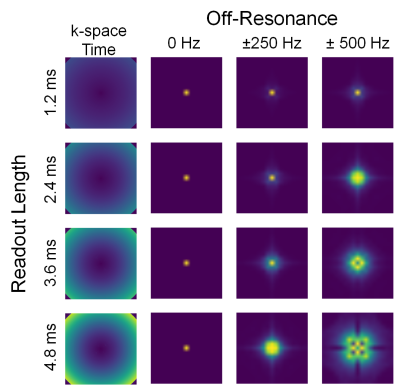
Training data was acquired on a 3T GE scanner with a 32-channel cardiac coil and a ferumoxytol-enhanced, ultra-short-echo-time (0.03ms) scan using 3D cones with short readouts between 0.9–1.5ms,2,3 reconstructed with ESPIRiT and no motion correction4. Multi-frequency autofocusing5 was used to correct the image for off-resonance and used as the reference image for supervised learning. Input data were generated by simulating the reference image at 101 off-resonances between [-500,500]Hz. Additionally, the off-resonance was applied across four different cones trajectories with [1.2,2.4,3.6,4.8]ms readouts to simulate a greater diversity of off-resonance PSFs (Figure 1). Each dataset was also divided into 64x64x64 patches to enable larger batch sizes. Eight datasets were used for a total of 400,000 training samples. Each voxel has a unique off-resonance PSF, thus every voxel can be effectively considered a separate data sample. For testing, 15 ferumoxytol-enhanced chest datasets with long readouts between 2.8–3.8ms were collected in addition to the short readouts. On average, the long-readout scans were 2.4x shorter than the short-readout scans.Deep Learning
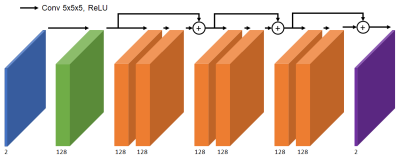
We used a supervised 3D convolutional neural network (CNN) to correct the off-resonance artifacts. The input to the network is a 3D image with two channels corresponding to real and imaginary components. The network architecture is three residual layers of 128 channels with 5x5x5 kernels6 (Figure 2). The output is the corrected image, trained with an L1 cost function using TensorFlow7.Result Analysis
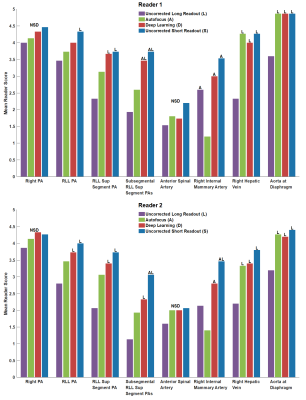
Two board-certified radiologists were independently presented in blinded fashion with four randomized, simultaneous images: uncorrected long readout, long readout with autofocus correction, long readout with deep learning correction, and uncorrected short readout. Image quality was evaluated for eight anatomic features, primarily for vessel definition, on a 5-point scale: 5-Excellent, 4-Good, 3-Moderate, 2-Poor, 1-Non-diagnostic. Significance of difference in scores (p<0.01) was determined by one-way ANOVA with post-hoc Tukey’s test8.Results
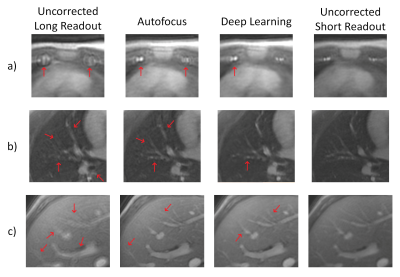
Sample images from each of the four methods are shown in Figure 3. The long-readout images have the most apparent off-resonance artifacts, and vessel definition is lost in Figure 3a) and b). Autofocus correction recovers vessel definition in the pulmonary artery and hepatic veins but the internal mammary arteries remain incoherent. Deep learning correction produces sharper pulmonary arteries and hepatic veins, producing longer coherent vessel segments. The internal mammary arteries are coherent and the left internal mammary arteries are distinguishable.
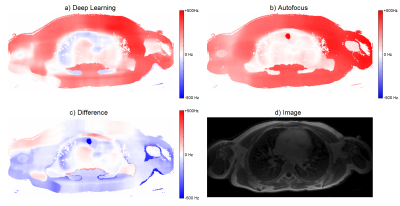
Field maps for both deep-learning-corrected and autofocus-corrected images were calculated by applying off-resonance on the original image and finding the closest match with the autofocus metric (Figure 4). The similarity of the field maps between the two methods gives confidence that the deep learning method is not hallucinating new structures into its output.
Statistical analysis results from two readers are shown in Figure 5. For both readers, deep learning images were not inferior (p<0.01) to any images in any features.
Discussion
These results demonstrate that the proposed deep learning method produces images non-inferior to short-readout images while having a 2.4x shorter scan. We demonstrated with a simple architecture that deep learning can effectively model and correct for off-resonance blurring. The performance can be further improved by longer training, more advanced architecture, and more accurate ground truth.
The deep learning images were also non-inferior to autofocus images and superior in several cases even though the neural network was trained on images corrected by autofocus. Although autofocus may not always resolve all off-resonance artifacts, perhaps the neural network is learning only the appropriate corrections.
Additionally, autofocus is computationally intensive because each candidate frequency must be simulated and reconstructed. Even with a field map, correction would take too long to be clinically viable. In contrast, our method does not need a field map and a typical dataset requires under a minute to be corrected with the proposed network, fast enough to be viable for clinical workflow.
From a theoretical approach, the signal equation for off-resonance without relaxation models off-resonance as a non-stationary convolution in the image domain1,9. Thus, it could be interpreted that the CNN is learning the appropriate non-stationary deconvolution kernel. An additional factor in increasing readout time is T2* decay and it is likely that the CNN is also learning to remove associated blur.
Acknowledgements
This research is supported by NIH R01 EB009690, NIH R01 HL127039, GE Healthcare, Joseph W. and Hon Mai Goodman Stanford Graduate Fellowship, and the National Science Foundation Graduate Research Fellowship under Grant No. DGE-114747.References
1. Chen W, Sica CT, Meyer CH. Fast conjugate phase image reconstruction based on a Chebyshev approximation to correct for B0 field inhomogeneity and concomitant gradients. Magn Reson Med. 2008;60(5):1104-1111.
2. Gurney PT, Hargreaves, BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med. 2006;55(3):575-582.
3. Carl M, Bydder GM, Du J. UTE imaging with simultaneous water and fat signal suppression using a time-efficient multispoke inversion recovery pulse sequence. Magn Reson Med. 2016:76(2):577-582.
4. Uecker M, et al. ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med. 2014;71(3):990-1001.
5. Noll DC, Pauly JM, Meyer CH, Nishimura DG, Macovski A. Deblurring for non-2D fourier transform magnetic resonance imaging. Magn Reson Med. 1992;25(2):319-333.
6. He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. Proc IEEE Conf Comp Vision Pattern Recognition. 2016.
7. Abadi M, et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. 2015.
8. McDonald J, Handbook of Biological Statistics. Baltimore: Sparky House Publishing; 2009.
9. Ahunbay E, Pipe JG. Rapid method for deblurring spiral MR images. Magn Reson Med. 2000;44(3):491-494.
Figures